Cancer Res Treat.
2016 Apr;48(2):698-707. 10.4143/crt.2015.118.
BRAF-Activated Long Noncoding RNA Modulates Papillary Thyroid Carcinoma Cell Proliferation through Regulating Thyroid Stimulating Hormone Receptor
- Affiliations
-
- 1Department of Surgery, Yantai Yuhuangding Hospital, Affiliated with Medical College of Qingdao University, Yantai, China. xujiedoc@126.com
- KMID: 2454348
- DOI: http://doi.org/10.4143/crt.2015.118
Abstract
- PURPOSE
The importance of long noncoding RNAs (lncRNAs) in tumorigenesis has recently been demonstrated. However, the role of lncRNAs in development of thyroid cancer remains largely unknown.
MATERIALS AND METHODS
Using quantitative reverse transcription polymerase chain reaction, expression of three lncRNAs, including BRAF-activated long noncoding RNA (BANCR), papillary thyroid cancer susceptibility candidate 3 (PTCSC3), and noncoding RNA associated with mitogen-activated protein kinase pathway and growth arrest (NAMA), was investigated in the current study.
RESULTS
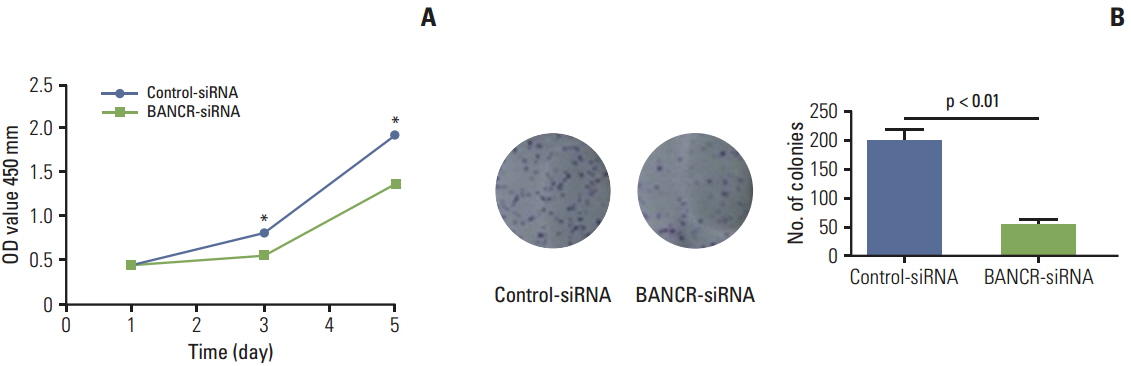
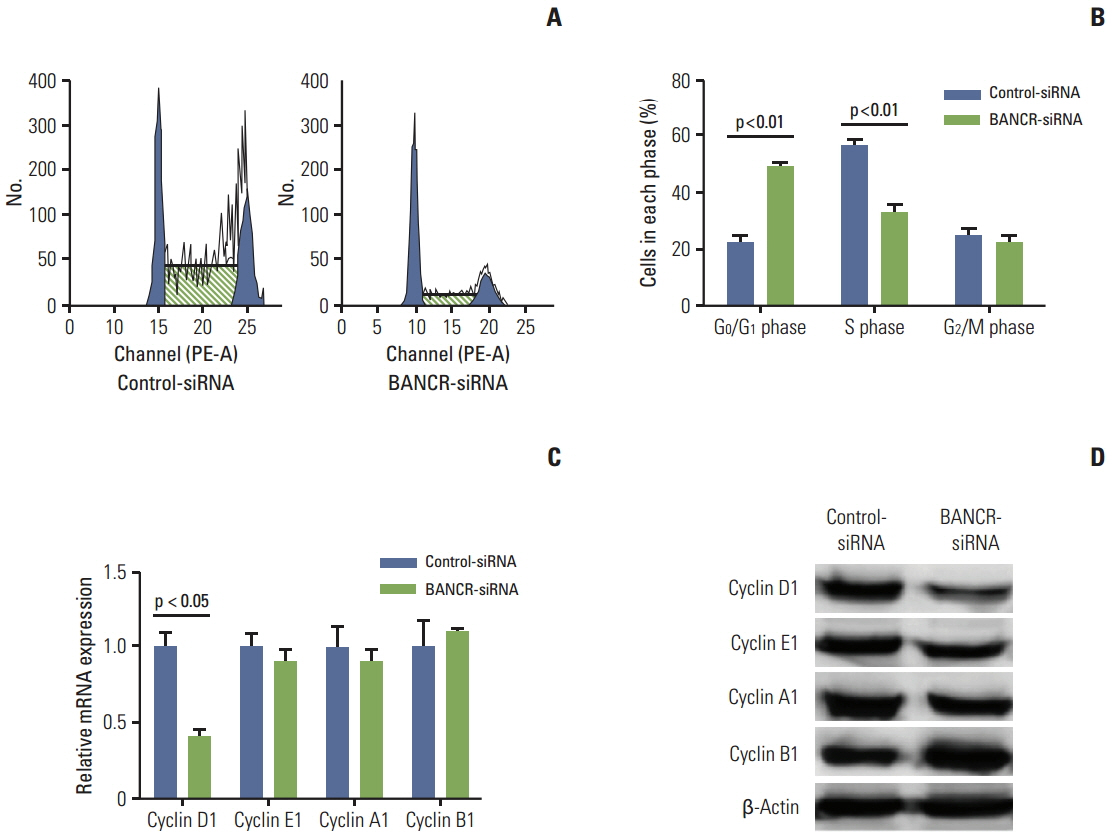
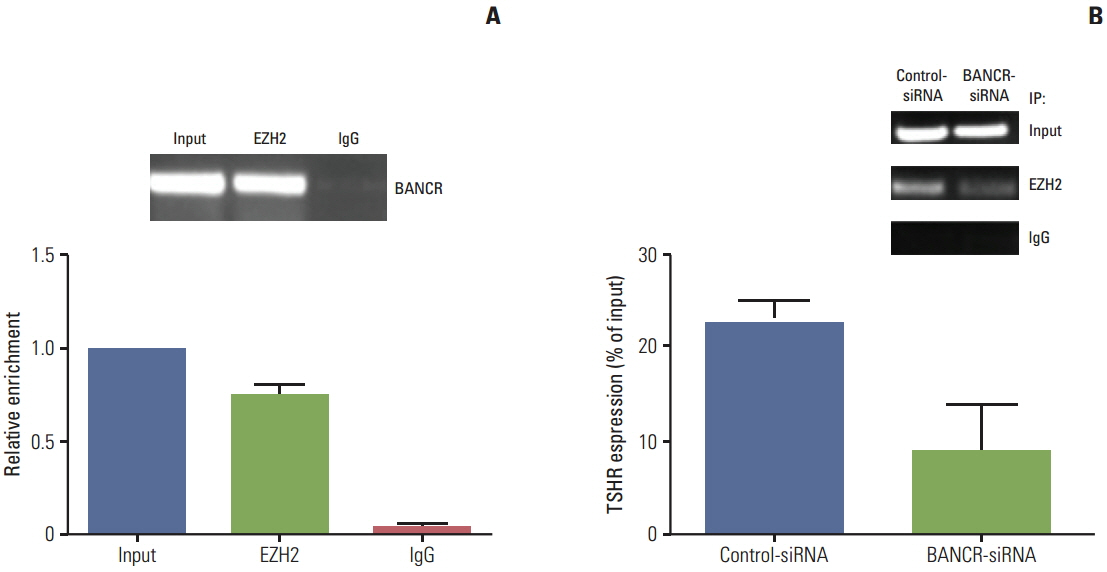
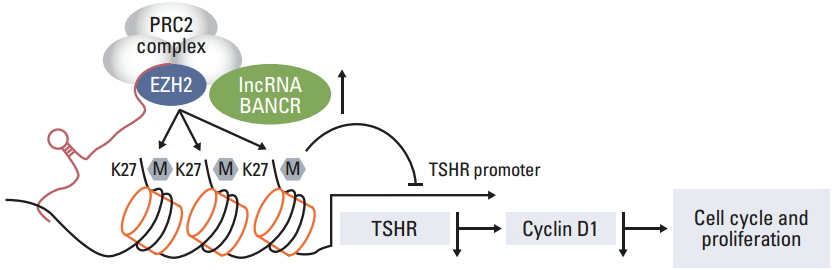
Of the three lncRNAs (BANCR, PTCSC3, and NAMA), expression of BANCR was significantly up-regulated while PTCSC3 and NAMA were significantly down-regulated in papillary thyroid carcinoma (PTC) compared to that in normal tissue. BANCR-knockdown in a PTC-derived cell line (IHH-4) resulted in significant suppression of thyroid stimulating hormone receptor (TSHR). BANCR-knockdown also led to inhibition of cell growth and cell cycle arrest at G0/G1 phase through down-regulation of cyclin D1. In addition, BANCR was enriched by polycomb enhancer of zeste homolog 2 (EZH2), and silencing BANCR led to decreased chromatin recruitment of EZH2, which resulted significantly reduced expression of TSHR.
CONCLUSION
These findings indicate that BANCR may contribute to the tumorigenesis of PTC through regulation of cyclin D1 and TSHR.
MeSH Terms
-
Carcinogenesis
Cell Cycle Checkpoints
Cell Line
Cell Proliferation*
Chromatin
Cyclin D1
Down-Regulation
Polymerase Chain Reaction
Protein Kinases
Receptors, Thyrotropin*
Reverse Transcription
RNA, Long Noncoding*
RNA, Untranslated
Thyroid Gland*
Thyroid Neoplasms*
Thyrotropin*
Chromatin
Cyclin D1
Protein Kinases
RNA, Long Noncoding
RNA, Untranslated
Receptors, Thyrotropin
Thyrotropin
Figure
Reference
-
References
1. Davies L, Welch HG. Increasing incidence of thyroid cancer in the United States, 1973-2002. JAMA. 2006; 295:2164–7.
Article2. Davies L, Welch HG. Current thyroid cancer trends in the United States. JAMA Otolaryngol Head Neck Surg. 2014; 140:317–22.
Article3. Wang Y, Guo Q, Zhao Y, Chen J, Wang S, Hu J, et al. BRAF-activated long non-coding RNA contributes to cell proliferation and activates autophagy in papillary thyroid carcinoma. Oncol Lett. 2014; 8:1947–52.
Article4. Fan M, Li X, Jiang W, Huang Y, Li J, Wang Z. A long non-coding RNA, PTCSC3, as a tumor suppressor and a target of miRNAs in thyroid cancer cells. Exp Ther Med. 2013; 5:1143–6.
Article5. Yoon H, He H, Nagy R, Davuluri R, Suster S, Schoenberg D, et al. Identification of a novel noncoding RNA gene, NAMA, that is downregulated in papillary thyroid carcinoma with BRAF mutation and associated with growth arrest. Int J Cancer. 2007; 121:767–75.6. Caretti G, Di Padova M, Micales B, Lyons GE, Sartorelli V. The Polycomb Ezh2 methyltransferase regulates muscle gene expression and skeletal muscle differentiation. Genes Dev. 2004; 18:2627–38.
Article7. Borbone E, Troncone G, Ferraro A, Jasencakova Z, Stojic L, Esposito F, et al. Enhancer of zeste homolog 2 overexpression has a role in the development of anaplastic thyroid carcinomas. J Clin Endocrinol Metab. 2011; 96:1029–38.
Article8. He H, Li W, Liyanarachch S, Jendrzejewski J, Srinivas M, Davuluri RV, et al. Genetic predisposition to papillary thyroid carcinoma: involvement of FOXE1, TSHR, and a novel lincRNA gene, PTCSC2. J Clin Endocrinol Metab. 2015; 100:E164–72.
Article9. Bufalo NE, Dos Santos RB, Marcello MA, Piai RP, Secolin R, Romaldini JH, et al. TSHR intronic polymorphisms (rs179247 and rs12885526) and their role in the susceptibility of the Brazilian population to Graves' disease and Graves' ophthalmopathy. J Endocrinol Invest. 2015; 38:555–61.
Article10. Khan MS, Pandith AA, Masoodi SR, Wani KA, Ul Hussain M, Mudassar S. Epigenetic silencing of TSHR gene in thyroid cancer patients in relation to their BRAF V600E mutation status. Endocrine. 2014; 47:449–55.
Article11. Huth S, Jaeschke H, Schaarschmidt J, Paschke R. Controversial constitutive TSHR activity: patients, physiology, and in vitro characterization. Horm Metab Res. 2014; 46:453–61.
Article12. Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006; 1:2315–9.
Article13. Telford WG, King LE, Fraker PJ. Rapid quantitation of apoptosis in pure and heterogeneous cell populations using flow cytometry. J Immunol Methods. 1994; 172:1–16.
Article14. Ngollo M, Lebert A, Dagdemir A, Judes G, Karsli-Ceppioglu S, Daures M, et al. The association between histone 3 lysine 27 trimethylation (H3K27me3) and prostate cancer: relationship with clinicopathological parameters. BMC Cancer. 2014; 14:994.
Article15. Esposito F, Tornincasa M, Pallante P, Federico A, Borbone E, Pierantoni GM, et al. Down-regulation of the miR-25 and miR-30d contributes to the development of anaplastic thyroid carcinoma targeting the polycomb protein EZH2. J Clin Endocrinol Metab. 2012; 97:E710–8.
Article16. Dong C, Hemminki K. Modification of cancer risks in offspring by sibling and parental cancers from 2,112,616 nuclear families. Int J Cancer. 2001; 92:144–50.
Article17. Mattick JS. Non-coding RNAs: the architects of eukaryotic complexity. EMBO Rep. 2001; 2:986–91.
Article18. Brosnan CA, Voinnet O. The long and the short of noncoding RNAs. Curr Opin Cell Biol. 2009; 21:416–25.
Article19. Ponting CP, Oliver PL, Reik W. Evolution and functions of long noncoding RNAs. Cell. 2009; 136:629–41.
Article20. Jia H, Osak M, Bogu GK, Stanton LW, Johnson R, Lipovich L. Genome-wide computational identification and manual annotation of human long noncoding RNA genes. RNA. 2010; 16:1478–87.
Article21. Flynn RA, Chang HY. Active chromatin and noncoding RNAs: an intimate relationship. Curr Opin Genet Dev. 2012; 22:172–8.
Article22. Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013; 193:651–69.
Article23. Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009; 10:155–9.
Article24. Varambally S, Cao Q, Mani RS, Shankar S, Wang X, Ateeq B, et al. Genomic loss of microRNA-101 leads to overexpression of histone methyltransferase EZH2 in cancer. Science. 2008; 322:1695–9.
Article25. Koh CM, Iwata T, Zheng Q, Bethel C, Yegnasubramanian S, De Marzo AM. Myc enforces overexpression of EZH2 in early prostatic neoplasia via transcriptional and post-transcriptional mechanisms. Oncotarget. 2011; 2:669–83.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expression of NF2 Modulates the Progression of BRAFV600E Mutated Thyroid Cancer Cells
- The Significance of TROP2 Expression in Predicting BRAF Mutations in Papillary Thyroid Carcinoma
- Medullary and Papillary Thyroid Carcinoma as a Collision Tumor: Report of Five Cases
- A Case of Multifocal Papillary Thyroid Carcinoma Consisting of One Encapsulated Follicular Variant with BRAF K601E Mutation and Three Conventional Types with BRAF V600E Mutation
- Osteopontin expression in papillary thyroid carcinoma and its relationship with the BRAF mutation and tumor characteristics