Lab Anim Res.
2011 Jun;27(2):109-116. 10.5625/lar.2011.27.2.109.
Isolation and Expression Profile of the Ca(2+)-Activated Chloride Channel-like Membrane Protein 6 Gene in Xenopus laevis
- Affiliations
-
- 1Department of Biochemistry and Molecular Cell Biology, College of Veterinary Medicine, Konkuk University, Seoul, Republic of Korea. jeongsm@konkuk.ac.kr
- 2Neuroscience Program, The University of Science and Technology, Seoul, Republic of Korea.
- 3Department of Physiology, Howard Hughes Medicine Institute, University of California, San Francisco, USA.
- 4The Institute for the 3Rs, College of Veterinary Medicine, Konkuk University, Seoul, Republic of Korea.
- 5College of Animal Bioscience and Technology, Konkuk University, Seoul, Republic of Korea.
- KMID: 2053659
- DOI: http://doi.org/10.5625/lar.2011.27.2.109
Abstract
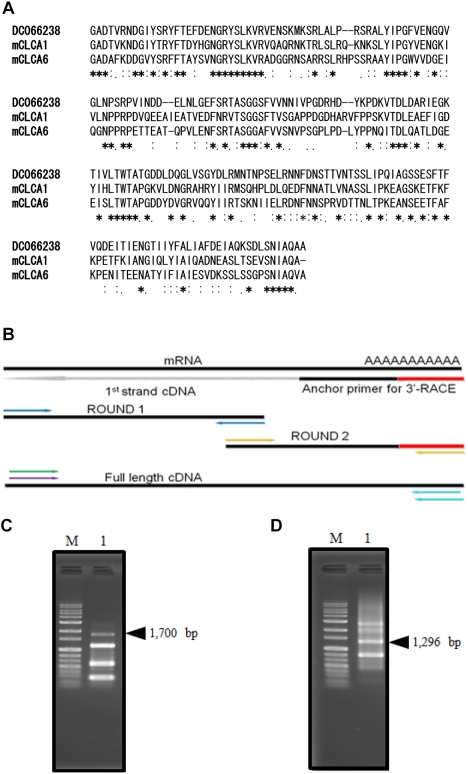
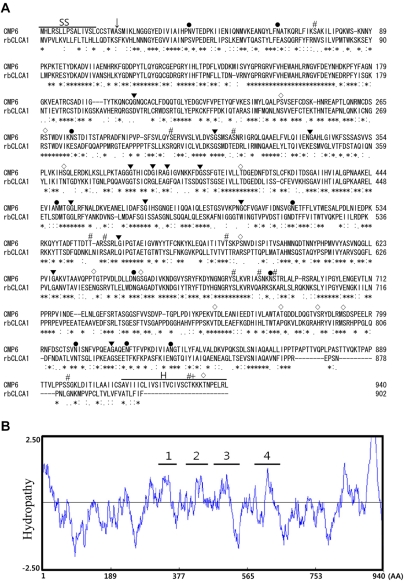
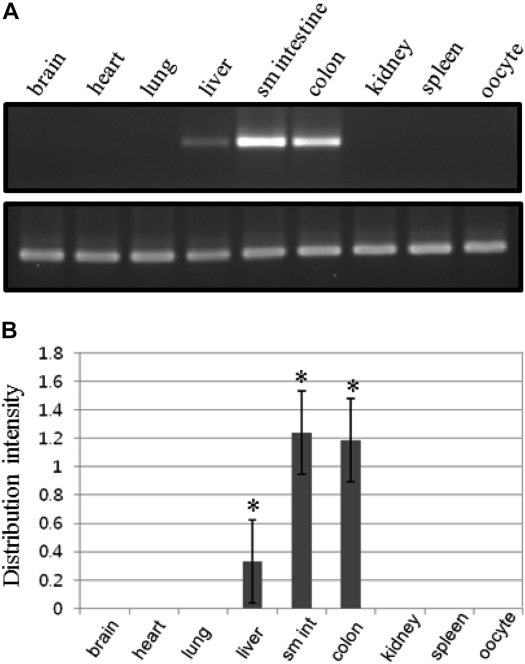
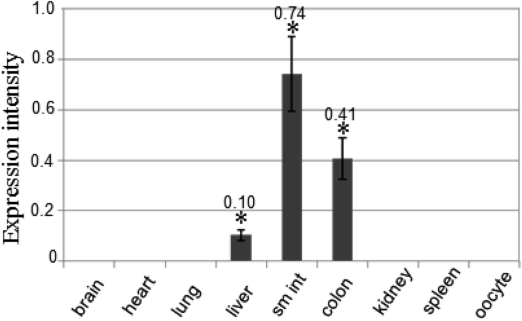
- To clone the first anion channel from Xenopus laevis (X. laevis), we isolated a calcium-activated chloride channel (CLCA)-like membrane protein 6 gene (CMP6) in X. laevis. As a first step in gene isolation, an expressed sequence tags database was screened to find the partial cDNA fragment. A putative partial cDNA sequence was obtained by comparison with rat CLCAs identified in our laboratory. First stranded cDNA was synthesized by reverse transcription polymerase-chain reaction (RT-PCR) using a specific primer designed for the target cDNA. Repeating the 5' and 3' rapid amplification of cDNA ends, full-length cDNA was constructed from the cDNA pool. The full-length CMP6 cDNA completed via 5'- and 3'-RACE was 2,940 bp long and had an open reading frame (ORF) of 940 amino acids. The predicted 940 polypeptides have four major transmembrane domains and showed about 50% identity with that of rat brain CLCAs in our previously published data. Semi-quantification analysis revealed that CMP6 was most abundantly expressed in small intestine, colon and liver. However, all tissues except small intestine, colon and liver had undetectable levels. This result became more credible after we did real-time PCR quantification for the target gene. In view of all CLCA studies focused on human or murine channels, this finding suggests a hypothetical protein as an ion channel, an X. laevis CLCA.
MeSH Terms
-
Amino Acids
Animals
Brain
Chloride Channels
Clone Cells
Colon
DNA, Complementary
Expressed Sequence Tags
Humans
Intestine, Small
Ion Channels
Liver
Membrane Proteins
Membranes
Open Reading Frames
Peptides
Rats
Real-Time Polymerase Chain Reaction
Resin Cements
Reverse Transcription
Staphylococcal Protein A
Tissue Distribution
Xenopus
Xenopus laevis
Amino Acids
Chloride Channels
DNA, Complementary
Ion Channels
Membrane Proteins
Peptides
Resin Cements
Staphylococcal Protein A
Figure
Reference
-
1. Kawai J, Shinagawa A, Shibata K, Yoshino M, Itoh M, Ishii Y, Arakawa T, Hara A, Fukunishi Y, Konno H, Adachi J, Fukuda S, Aizawa K, Izawa M, Nishi K, Kiyosawa H, Kondo S, Yamanaka I, Saito T, Okazaki Y, Gojobori T, Bono H, Kasukawa T, Saito R, Kadota K, Matsuda H, Ashburner M, Batalov S, Casavant T, Fleischmann W, Gaasterland T, Gissi C, King B, Kochiwa H, Kuehl P, Lewis S, Matsuo Y, Nikaido I, Pesole G, Quackenbush J, Schriml LM, Staubli F, Suzuki R, Tomita M, Wagner L, Washio T, Sakai K, Okido T, Furuno M, Aono H, Baldarelli R, Barsh G, Blake J, Boffelli D, Bojunga N, Carninci P, de Bonaldo MF, Brownstein MJ, Bult C, Fletcher C, Fujita M, Gariboldi M, Gustincich S, Hill D, Hofmann M, Hume DA, Kamiya M, Lee NH, Lyons P, Marchionni L, Mashima J, Mazzarelli J, Mombaerts P, Nordone P, Ring B, Ringwald M, Rodriguez I, Sakamoto N, Sasaki H, Sato K, Schönbach C, Seya T, Shibata Y, Storch KF, Suzuki H, Toyooka K, Wang KH, Weitz C, Whittaker C, Wilming L, Wynshaw-Boris A, Yoshida K, Hasegawa Y, Kawaji H, Kohtsuki S, Hayashizaki Y. RIKEN Genome Exploration Research Group Phase II Team and the FANTOM Consortium. Functional annotation of a full-length mouse cDNA collection. Nature. 2001; 409:685–690. PMID: 11217851.2. Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, Scott G, Steffen D, Worley KC, Burch PE, Okwuonu G, Hines S, Lewis L, DeRamo C, Delgado O, Dugan-Rocha S, Miner G, Morgan M, Hawes A, Gill R, Celera , Holt RA, Adams MD, Amanatides PG, Baden-Tillson H, Barnstead M, Chin S, Evans CA, Ferriera S, Fosler C, Glodek A, Gu Z, Jennings D, Kraft CL, Nguyen T, Pfannkoch CM, Sitter C, Sutton GG, Venter JC, Woodage T, Smith D, Lee HM, Gustafson E, Cahill P, Kana A, Doucette-Stamm L, Weinstock K, Fechtel K, Weiss RB, Dunn DM, Green ED, Blakesley RW, Bouffard GG, De Jong PJ, Osoegawa K, Zhu B, Marra M, Schein J, Bosdet I, Fjell C, Jones S, Krzywinski M, Mathewson C, Siddiqui A, Wye N, McPherson J, Zhao S, Fraser CM, Shetty J, Shatsman S, Geer K, Chen Y, Abramzon S, Nierman WC, Havlak PH, Chen R, Durbin KJ, Egan A, Ren Y, Song XZ, Li B, Liu Y, Qin X, Cawley S, Worley KC, Cooney AJ, D'Souza LM, Martin K, Wu JQ, Gonzalez-Garay ML, Jackson AR, Kalafus KJ, McLeod MP, Milosavljevic A, Virk D, Volkov A, Wheeler DA, Zhang Z, Bailey JA, Eichler EE, Tuzun E, Birney E, Mongin E, Ureta-Vidal A, Woodwark C, Zdobnov E, Bork P, Suyama M, Torrents D, Alexandersson M, Trask BJ, Young JM, Huang H, Wang H, Xing H, Daniels S, Gietzen D, Schmidt J, Stevens K, Vitt U, Wingrove J, Camara F, Mar Albà M, Abril JF, Guigo R, Smit A, Dubchak I, Rubin EM, Couronne O, Poliakov A, Hübner N, Ganten D, Goesele C, Hummel O, Kreitler T, Lee YA, Monti J, Schulz H, Zimdahl H, Himmelbauer H, Lehrach H, Jacob HJ, Bromberg S, Gullings-Handley J, Jensen-Seaman MI, Kwitek AE, Lazar J, Pasko D, Tonellato PJ, Twigger S, Ponting CP, Duarte JM, Rice S, Goodstadt L, Beatson SA, Emes RD, Winter EE, Webber C, Brandt P, Nyakatura G, Adetobi M, Chiaromonte F, Elnitski L, Eswara P, Hardison RC, Hou M, Kolbe D, Makova K, Miller W, Nekrutenko A, Riemer C, Schwartz S, Taylor J, Yang S, Zhang Y, Lindpaintner K, Andrews TD, Caccamo M, Clamp M, Clarke L, Curwen V, Durbin R, Eyras E, Searle SM, Cooper GM, Batzoglou S, Brudno M, Sidow A, Stone EA, Venter JC, Payseur BA, Bourque G, López-Otín C, Puente XS, Chakrabarti K, Chatterji S, Dewey C, Pachter L, Bray N, Yap VB, Caspi A, Tesler G, Pevzner PA, Haussler D, Roskin KM, Baertsch R, Clawson H, Furey TS, Hinrichs AS, Karolchik D, Kent WJ, Rosenbloom KR, Trumbower H, Weirauch M, Cooper DN, Stenson PD, Ma B, Brent M, Arumugam M, Shteynberg D, Copley RR, Taylor MS, Riethman H, Mudunuri U, Peterson J, Guyer M, Felsenfeld A, Old S, Mockrin S, Collins F. Rat Genome Sequencing Project Consortium. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004; 428:493–521. PMID: 15057822.3. Hartzell CH, Putzier I, Arreola J. Calcium-activated chloride channels. Annu Rev Physiol. 2005; 67:719–758. PMID: 15709976.
Article4. Loewen ME, Forsyth GW. Structure and function of CLCA proteins. Physiol Rev. 2005; 85:1061–1092. PMID: 15987802.
Article5. Caputo A, Caci E, Ferrera L, Pedemonte N, Barsanti C, Sondo E, Pfeffer U, Ravazzolo R, Zegarra-Moran O, Galietta LJ. TMEM16A, a membrane protein associated with calcium-dependent chloride channel activity. Science. 2008; 322:590–594. PMID: 18772398.
Article6. Huang F, Rock JR, Harfe BD, Cheng T, Huang X, Jan YN, Jan LY. Studies on expression and function of the TMEM16A calcium-activated chloride channel. Proc Natl Acad Sci USA. 2009; 106:21413–21418. PMID: 19965375.
Article7. Yang YD, Cho H, Koo JY, Tak MH, Cho Y, Shin WS, Park SP, Lee J, Lee B, Kim BM, Raouf R, Shin YK, Oh U. TMEM16A confers receptor-activated calcium-dependent chloride conductance. Nature. 2008; 455:1210–1215. PMID: 18724360.
Article8. Schroeder BC, Cheng T, Jan YN, Jan LY. Expression cloning of TMEM16A as a calcium-activated chloride channel subunit. Cell. 2008; 134:1019–1029. PMID: 18805094.
Article9. Ryu RH, Oh SJ, Lee RM, Jeong SW, Jan LY, Lee CH, Lee CJ, Jeong SM. Cloning and heterologous expression of new xANO2 from Xenopus laevis. Biochem Biophys Res Commun. 2011; 408(4):559–565. PMID: 21521635.10. Cunningham SA, Awayda MS, Bubien JK, Ismailov II, Arrate MP, Berdiev BK, Benos DJ, Fuller CM. Cloning of an epithelial chloride channel from bovine trachea. J Biol Chem. 1995; 270:31016–31026. PMID: 8537359.
Article11. Evans SR, Thoreson WB, Beck CL. Molecular and functional analyses of two new calcium-activated chloride channel family members from mouse eye and intestine. J Biol Chem. 2004; 279:41792–41800. PMID: 15284223.
Article12. Lee D, Ha S, Kho Y, Kim J, Cho K, Baik M, Choi Y. Induction of mouse Ca(2+)-sensitive chloride channel 2 gene during involution of mammary gland. Biochem Biophys Res Commun. 1999; 264:933–937. PMID: 10544033.
Article13. Agnel M, Vermat T, Culouscou JM. Identification of three novel members of the calcium dependent chloride channel (CaCC) family predominantly expressed in the digestive tract and trachea. FEBS Lett. 1999; 455:295–301. PMID: 10437792.14. Gaspar KJ, Racette KJ, Gordon JR, Loewen ME, Forsyth GW. Cloning a chloride conductance mediator from the apical membrane of porcine ileal enterocytes. Physiol Genomics. 2000; 3:101–111. PMID: 11015605.
Article15. Jeong SM, Park HK, Yoon IS, Lee JH, Kim JH, Jang CG, Lee CJ, Nah SY. Cloning and expression of Ca2+-activated chloride channel from rat brain. Biochem Biophys Res Commun. 2005; 334:569–576. PMID: 16023076.16. Yoon IS, Jeong SM, Lee SN, Lee JH, Kim JH, Pyo MK, Lee BH, Choi SH, Rhim H, Choe H, Nah SY. Cloning and heterologous expression of a Ca2+-activated chloride channel isoform from rat brain. Biol Pharm Bull. 2006; 29:2168–2173. PMID: 17077509.
Article17. Gruber AD, Pauli BU. Tumorigenicity of human breast cancer is associated with loss of the Ca2+-activated chloride channel CLCA2. Cancer Res. 1999; 59:5488–5491. PMID: 10554024.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Cloning of Xenopus laevis TRPV2 by Gene Prediction
- Na, K-ATPase beta2 isoform (atp1b2) expressed in the retina of Xenopus
- Study on histological features and Bmp4 expression pattern during tooth formation and replacement in Xenopus laevis
- Molecular characterisation of pancreatic zymogen granule ion channel and regulator proteins involved in exocytosis
- Temporal Inhibition of FGF Signal on Endoderm Formation during Early Xenopus laevis Embryogenesis