J Breast Cancer.
2016 Dec;19(4):385-393. 10.4048/jbc.2016.19.4.385.
Expression of T-Lymphocyte Markers in Human Epidermal Growth Factor Receptor 2-Positive Breast Cancer
- Affiliations
-
- 1Department of Surgery, Yonsei University College of Medicine, Seoul, Korea. psh1025@yuhs.ac
- 2Frontier Research Institute of Convergence Sports Science, Yonsei University, Seoul, Korea.
- KMID: 2362924
- DOI: http://doi.org/10.4048/jbc.2016.19.4.385
Abstract
- PURPOSE
The present study aimed to examine the clinical implications of CD4, CD8, and FOXP3 expression on the prognosis of human epidermal growth factor receptor 2 (HER2)-positive breast cancer using a web-based database, and to compare the immunohistochemical expression of T-lymphocyte markers using primary and metastatic HER2-positive tumor tissues before and after HER2-targeted therapy.
METHODS
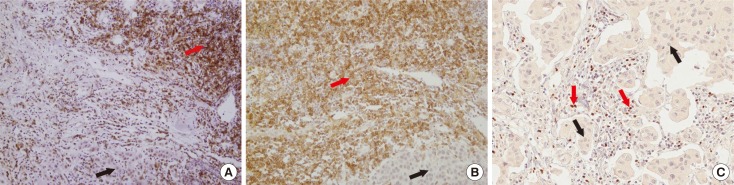
Using the cBioPortal for Cancer Genomics and Kaplan-Meier plotter, the mRNA expression, association between T-lymphocyte markers, and survival in HER2-positive cancers were investigated according to various cutoff levels. Immunohistochemistry analysis was performed using paired primary and metastatic tissues of 29 HER2-positive tumors treated with systemic chemotherapy and HER2-directed therapy.
RESULTS
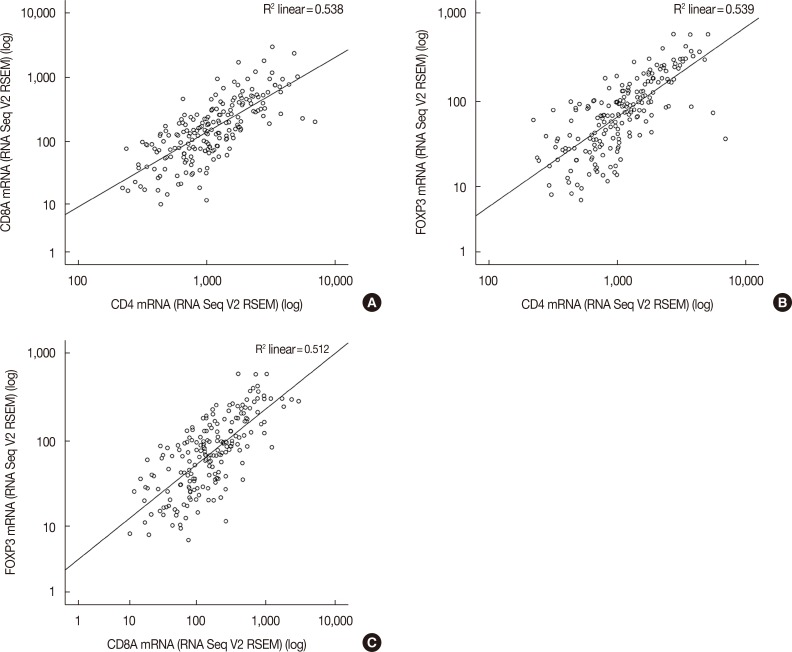
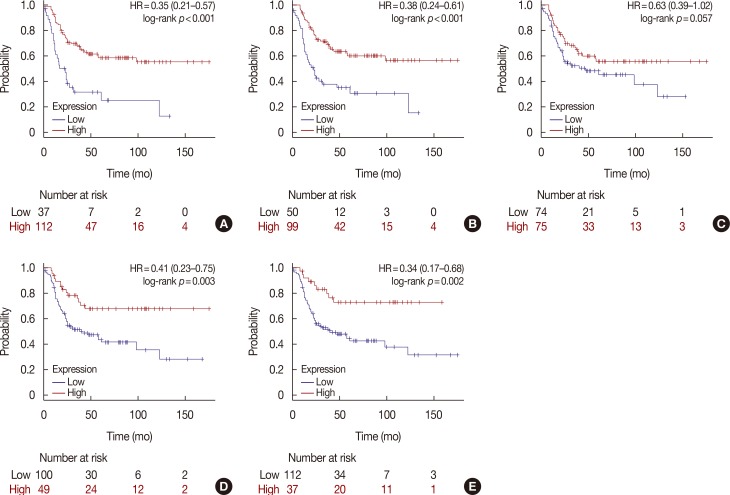
HER2 mRNA was mutually exclusive of T-lymphocyte markers, and a significant correlation between T-cell markers was observed in the cBioPortal for Cancer Genomics. According to analysis of the Kaplan-Meier plotter, the impact of T-lymphocyte marker expression on survival was statistically insignificant in clinical HER2-positive tumors, irrespective of the cutoff levels. However, in the intrinsic HER2-positive subtype, the individual analyses of T-cell markers except for FOXP3 and combined analysis showed significantly favorable survival irrespective of cutoff points. Although the small clinical sample size made it difficult to show the statistical relevance of immunohistochemistry findings, good responses to neoadjuvant treatments might be associated with positive expression of combined T-lymphocyte markers, and approximately half of the samples showed discordance of combined markers between baseline and resistant tumors.
CONCLUSION
T-lymphocyte markers could be favorable prognostic factors in HER2-positive breast cancers; however, a consensus on patient section criteria, detection methods, and cutoff value could not be reached. The resistance to HER2-directed therapy might involve different and personalized mechanisms, and further research is required to understand the association between immune function and HER2 expression and to overcome the resistance mechanisms to HER2-targeted therapies.
MeSH Terms
-
Biomarkers
Breast
Breast Neoplasms
Consensus
Drug Resistance
Drug Therapy
Epidermal Growth Factor*
Genomics
Humans*
Immunohistochemistry
Neoadjuvant Therapy
Prognosis
Receptor, Epidermal Growth Factor*
RNA, Messenger
Sample Size
T-Lymphocytes*
Biomarkers
Epidermal Growth Factor
RNA, Messenger
Receptor, Epidermal Growth Factor
Figure
Reference
-
1. Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survival with amplification of the HER-2/neu oncogene. Science. 1987; 235:177–182. PMID: 3798106.2. Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001; 344:783–792. PMID: 11248153.
Article3. Romond EH, Perez EA, Bryant J, Suman VJ, Geyer CE Jr, Davidson NE, et al. Trastuzumab plus adjuvant chemotherapy for operable HER2-positive breast cancer. N Engl J Med. 2005; 353:1673–1684. PMID: 16236738.
Article4. Santa-Maria CA, Nye L, Mutonga MB, Jain S, Gradishar WJ. Management of metastatic HER2-positive breast cancer: where are we and where do we go from here? Oncology (Williston Park). 2016; 30:148–155. PMID: 26892151.5. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011; 144:646–674. PMID: 21376230.
Article6. Vu T, Claret FX. Trastuzumab: updated mechanisms of action and resistance in breast cancer. Front Oncol. 2012; 2:62. PMID: 22720269.
Article7. Luque-Cabal M, García-Teijido P, Fernández-Pérez Y, Sánchez-Lorenzo L, Palacio-Vázquez I. Mechanisms behind the resistance to trastuzumab in HER2-amplified breast cancer and strategies to overcome it. Clin Med Insights Oncol. 2016; 10(Suppl 1):21–30. PMID: 27042153.
Article8. Datta J, Berk E, Xu S, Fitzpatrick E, Rosemblit C, Lowenfeld L, et al. Anti-HER2 CD4(+) T-helper type 1 response is a novel immune correlate to pathologic response following neoadjuvant therapy in HER2-positive breast cancer. Breast Cancer Res. 2015; 17:71. PMID: 25997452.
Article9. Asano Y, Kashiwagi S, Goto W, Kurata K, Noda S, Takashima T, et al. Tumour-infiltrating CD8 to FOXP3 lymphocyte ratio in predicting treatment responses to neoadjuvant chemotherapy of aggressive breast cancer. Br J Surg. 2016; 103:845–854. PMID: 26953091.
Article10. Lee HJ, Seo JY, Ahn JH, Ahn SH, Gong G. Tumor-associated lymphocytes predict response to neoadjuvant chemotherapy in breast cancer patients. J Breast Cancer. 2013; 16:32–39. PMID: 23593079.
Article11. Ali HR, Provenzano E, Dawson SJ, Blows FM, Liu B, Shah M, et al. Association between CD8+T-cell infiltration and breast cancer survival in 12,439 patients. Ann Oncol. 2014; 25:1536–1543. PMID: 24915873.12. Loi S, Michiels S, Salgado R, Sirtaine N, Jose V, Fumagalli D, et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: results from the FinHER trial. Ann Oncol. 2014; 25:1544–1550. PMID: 24608200.
Article13. Ahn SG, Jeong J, Hong S, Jung WH. Current issues and clinical evidence in tumor-infiltrating lymphocytes in breast cancer. J Pathol Transl Med. 2015; 49:355–363. PMID: 26278518.
Article14. Salgado R, Denkert C, Demaria S, Sirtaine N, Klauschen F, Pruneri G, et al. The evaluation of tumor-infiltrating lymphocytes (TILs) in breast cancer: recommendations by an International TILs Working Group 2014. Ann Oncol. 2015; 26:259–271. PMID: 25214542.
Article15. Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012; 2:401–404. PMID: 22588877.16. Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013; 6:pl1. PMID: 23550210.
Article17. Györffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat. 2010; 123:725–731. PMID: 20020197.
Article18. Park S, Ko YS, Koo JS, Sohn J, Kim SI, Park BW. Expression of growth factor receptor family before and after targeted therapy in human epidermal growth factor receptor-2 positive breast cancer tissues. Korean J Clin Oncol. 2015; 11:12–19.
Article19. Hammond ME, Hayes DF, Dowsett M, Allred DC, Hagerty KL, Badve S, et al. American Society of Clinical Oncology/College of American Pathologists guideline recommendations for immunohistochemical testing of estrogen and progesterone receptors in breast cancer. J Clin Oncol. 2010; 28:2784–2795. PMID: 20404251.
Article20. Wolff AC, Hammond ME, Hicks DG, Dowsett M, McShane LM, Allison KH, et al. Recommendations for human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline update. J Clin Oncol. 2013; 31:3997–4013. PMID: 24101045.
Article21. Stanton SE, Adams S, Disis ML. Variation in the Incidence and magnitude of tumor-infiltrating lymphocytes in breast cancer subtypes: a systematic review. JAMA Oncol. 2016; 2:1354–1360. PMID: 27355489.22. Wang K, Xu J, Zhang T, Xue D. Tumor-infiltrating lymphocytes in breast cancer predict the response to chemotherapy and survival outcome: a meta-analysis. Oncotarget. 2016; 7:44288–44298. PMID: 27329588.
Article23. Kim S, Lee A, Lim W, Park S, Cho MS, Koo H, et al. Zonal difference and prognostic significance of foxp3 regulatory T cell infiltration in breast cancer. J Breast Cancer. 2014; 17:8–17. PMID: 24744792.
Article24. Denkert C, Wienert S, Poterie A, Loibl S, Budczies J, Badve S, et al. Standardized evaluation of tumor-infiltrating lymphocytes in breast cancer: results of the ring studies of the international immuno-oncology biomarker working group. Mod Pathol. 2016; 29:1155–1164. PMID: 27363491.
Article25. Perez EA, Thompson EA, Ballman KV, Anderson SK, Asmann YW, Kalari KR, et al. Genomic analysis reveals that immune function genes are strongly linked to clinical outcome in the North Central Cancer Treatment Group n9831 Adjuvant Trastuzumab Trial. J Clin Oncol. 2015; 33:701–708. PMID: 25605861.
Article26. Gagliato DM, Jardim DL, Marchesi MS, Hortobagyi GN. Mechanisms of resistance and sensitivity to anti-HER2 therapies in HER2+ breast cancer. Oncotarget. 2016; 7:64431–64446.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prognostic significance of epidermal growth factor receptor expression in human gastric carcinoma
- The Prognostic Value of Epidermal Growth Factor Receptor in Primary Breast Cancer
- Human Epidermal Growth Factor Receptor 2-positive Mucinous Carcinoma with Signet Ring Cell Differentiation, Which Showed Complete Response after Neoadjuvant Chemotherapy
- Amplification of epidermal growth factor receptor gene in primary cervical cancer
- Amplification of epidermal growth factor receptor gene in primary cervical cancer