Korean J Physiol Pharmacol.
2019 Sep;23(5):367-379. 10.4196/kjpp.2019.23.5.367.
Profiling of remote skeletal muscle gene changes resulting from stimulation of atopic dermatitis disease in NC/Nga mouse model
- Affiliations
-
- 1Department of Physiology, Chung-Ang University College of Medicine, Seoul 06974, Korea. akdongyi01@cau.ac.kr
- 2Department of Medicine, Chung-Ang University College of Medicine, Seoul 06974, Korea.
- 3Department of Family Medicine, Chung-Ang University Hospital, Chung-Ang University College of Medicine, Seoul 06973, Korea. girlpower219@cau.ac.kr
- KMID: 2455813
- DOI: http://doi.org/10.4196/kjpp.2019.23.5.367
Abstract
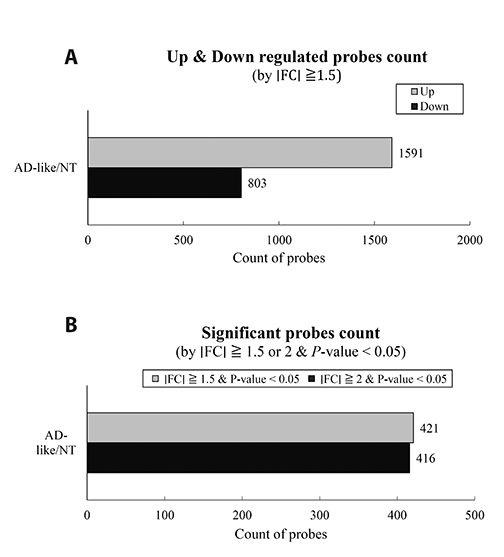
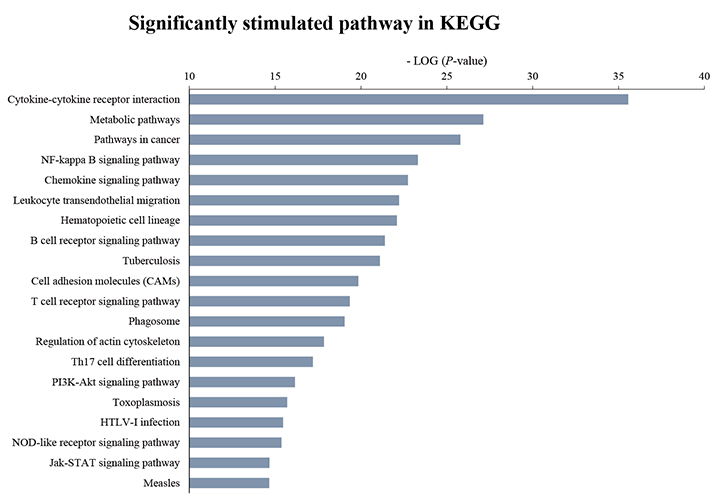
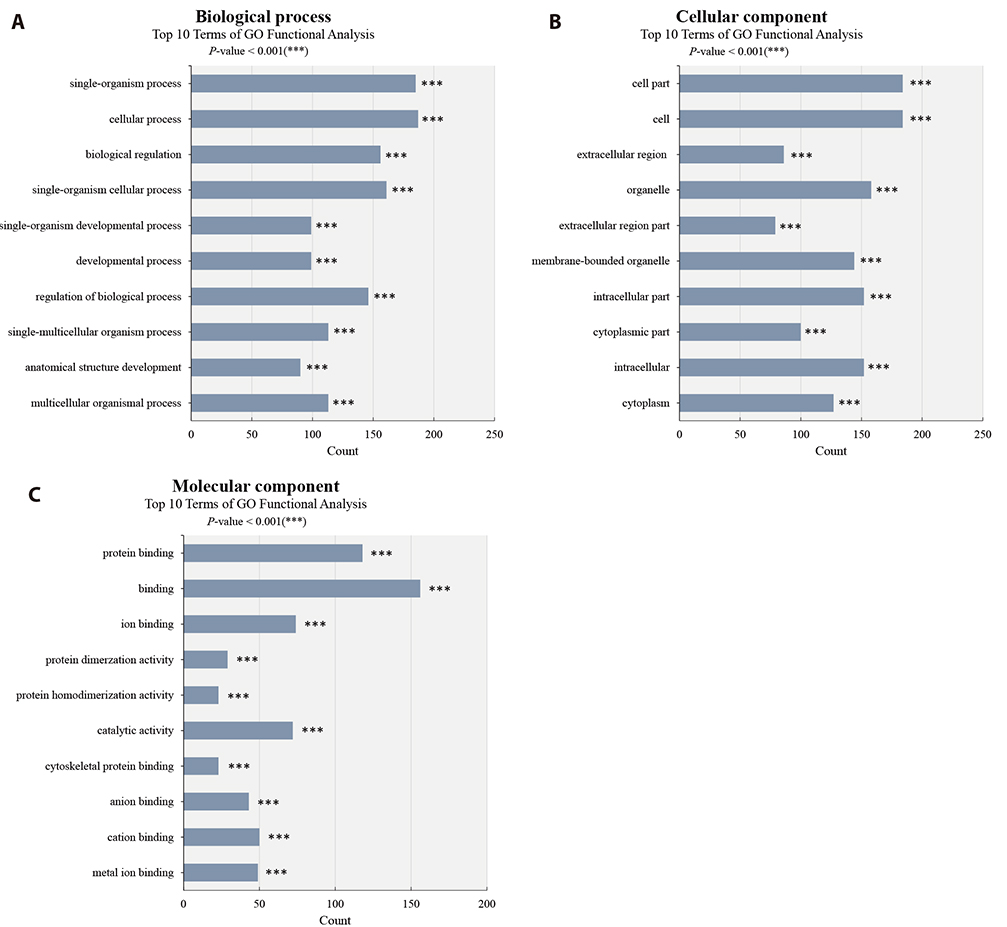
- Although atopic dermatitis (AD) is known to be a representative skin disorder, it also affects the systemic immune response. In a recent study, myoblasts were shown to be involved in the immune regulation, but the roles of muscle cells in AD are poorly understood. We aimed to identify the relationship between mitochondria and atopy by genome-wide analysis of skeletal muscles in mice. We induced AD-like symptoms using house dust mite (HDM) extract in NC/Nga mice. The transcriptional profiles of the untreated group and HDM-induced AD-like group were analyzed and compared using microarray, differentially expressed gene and functional pathway analyses, and protein interaction network construction. Our microarray analysis demonstrated that immune response-, calcium handling-, and mitochondrial metabolism-related genes were differentially expressed. In the Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology pathway analyses, immune response pathways involved in cytokine interaction, nuclear factor-kappa B, and T-cell receptor signaling, calcium handling pathways, and mitochondria metabolism pathways involved in the citrate cycle were significantly upregulated. In protein interaction network analysis, chemokine family-, muscle contraction process-, and immune response-related genes were identified as hub genes with many interactions. In addition, mitochondrial pathways involved in calcium signaling, cardiac muscle contraction, tricarboxylic acid cycle, oxidation-reduction process, and calcium-mediated signaling were significantly stimulated in KEGG and Gene Ontology analyses. Our results provide a comprehensive understanding of the genome-wide transcriptional changes of HDM-induced AD-like symptoms and the indicated genes that could be used as AD clinical biomarkers.
MeSH Terms
-
Animals
Biomarkers
Calcium
Calcium Signaling
Citric Acid
Citric Acid Cycle
Cytokines
Dermatitis, Atopic*
Gene Ontology
Genome
Metabolism
Mice*
Microarray Analysis
Mitochondria
Muscle Cells
Muscle Contraction
Muscle, Skeletal*
Myoblasts
Myocardium
Oxidation-Reduction
Protein Interaction Maps
Pyroglyphidae
Receptors, Antigen, T-Cell
Skin
Biomarkers
Calcium
Citric Acid
Cytokines
Receptors, Antigen, T-Cell
Figure
Reference
-
1. Yosipovitch G, Greaves MW, Schmelz M. Itch. Lancet. 2003; 361:690–694.
Article2. Williams HC. Epidemiology of atopic dermatitis. Clin Exp Dermatol. 2000; 25:522–529.
Article3. Weidinger S, Novak N. Atopic dermatitis. Lancet. 2016; 387:1109–1122.
Article4. Ikoma A, Steinhoff M, Ständer S, Yosipovitch G, Schmelz M. The neurobiology of itch. Nat Rev Neurosci. 2006; 7:535–547.
Article5. Goebels N, Michaelis D, Wekerle H, Hohlfeld R. Human myoblasts as antigen-presenting cells. J Immunol. 1992; 149:661–667.6. Curnow J, Corlett L, Willcox N, Vincent A. Presentation by myoblasts of an epitope from endogenous acetylcholine receptor indicates a potential role in the spreading of the immune response. J Neuroimmunol. 2001; 115:127–134.
Article7. Gallucci S, Provenzano C, Mazzarelli P, Scuderi F, Bartoccioni E. Myoblasts produce IL-6 in response to inflammatory stimuli. Int Immunol. 1998; 10:267–273.
Article8. Nagaraju K. Immunological capabilities of skeletal muscle cells. Acta Physiol Scand. 2001; 171:215–223.
Article9. Wiendl H, Mitsdoerffer M, Schneider D, Chen L, Lochmüller H, Melms A, Weller M. Human muscle cells express a B7-related molecule, B7-H1, with strong negative immune regulatory potential: a novel mechanism of counterbalancing the immune attack in idiopathic inflammatory myopathies. FASEB J. 2003; 17:1892–1894.
Article10. Koziol-White CJ, Panettieri RA Jr. Airway smooth muscle and immunomodulation in acute exacerbations of airway disease. Immunol Rev. 2011; 242:178–185.
Article11. Damera G, Tliba O, Panettieri RA Jr. Airway smooth muscle as an immunomodulatory cell. Pulm Pharmacol Ther. 2009; 22:353–359.
Article12. Aguilera-Aguirre L, Bacsi A, Saavedra-Molina A, Kurosky A, Sur S, Boldogh I. Mitochondrial dysfunction increases allergic airway inflammation. J Immunol. 2009; 183:5379–5387.
Article13. West AP, Shadel GS, Ghosh S. Mitochondria in innate immune responses. Nat Rev Immunol. 2011; 11:389–402.
Article14. Trian T, Benard G, Begueret H, Rossignol R, Girodet PO, Ghosh D, Ousova O, Vernejoux JM, Marthan R, Tunon-de-Lara JM, Berger P. Bronchial smooth muscle remodeling involves calcium-dependent enhanced mitochondrial biogenesis in asthma. J Exp Med. 2007; 204:3173–3181.
Article15. Girodet PO, Allard B, Thumerel M, Begueret H, Dupin I, Ousova O, Lassalle R, Maurat E, Ozier A, Trian T, Marthan R, Berger P. Bronchial smooth muscle remodeling in nonsevere asthma. Am J Respir Crit Care Med. 2016; 193:627–633.
Article16. Xu YD, Cui JM, Wang Y, Yin LM, Gao CK, Liu YY, Yang YQ. The early asthmatic response is associated with glycolysis, calcium binding and mitochondria activity as revealed by proteomic analysis in rats. Respir Res. 2010; 11:107.
Article17. Bunyavanich S, Schadt EE, Himes BE, Lasky-Su J, Qiu W, Lazarus R, Ziniti JP, Cohain A, Linderman M, Torgerson DG, Eng CS, Pino-Yanes M, Padhukasahasram B, Yang JJ, Mathias RA, Beaty TH, Li X, Graves P, Romieu I, Navarro Bdel R, et al. Integrated genome-wide association, coexpression network, and expression single nucleotide polymorphism analysis identifies novel pathway in allergic rhinitis. BMC Med Genomics. 2014; 7:48.
Article18. Kay AB. Allergy and allergic diseases. First of two parts. N Engl J Med. 2001; 344:30–37.19. Raby BA, Klanderman B, Murphy A, Mazza S, Camargo CA Jr, Silverman EK, Weiss ST. A common mitochondrial haplogroup is associated with elevated total serum IgE levels. J Allergy Clin Immunol. 2007; 120:351–358.
Article20. Bradley M, Kockum I, Söderhäll C, Van Hage-Hamsten M, Luthman H, Nordenskjöld M, Wahlgren CF. Characterization by phenotype of families with atopic dermatitis. Acta Derm Venereol. 2000; 80:106–110.21. Dold S, Wjst M, von Mutius E, Reitmeir P, Stiepel E. Genetic risk for asthma, allergic rhinitis, and atopic dermatitis. Arch Dis Child. 1992; 67:1018–1022.
Article22. Morar N, Willis-Owen SA, Moffatt MF, Cookson WO. The genetics of atopic dermatitis. J Allergy Clin Immunol. 2006; 118:24–34.
Article23. Iyer D, Mishra N, Agrawal A. Mitochondrial function in allergic disease. Curr Allergy Asthma Rep. 2017; 17:29.
Article24. Spergel JM, Paller AS. Atopic dermatitis and the atopic march. J Allergy Clin Immunol. 2003; 112(6 Suppl):S118–S127.
Article25. Čepelak I, Dodig S, Pavić I. Filaggrin and atopic march. Biochem Med (Zagreb). 2019; 29:020501.
Article26. Guo W, Jiang L, Bhasin S, Khan SM, Swerdlow RH. DNA extraction procedures meaningfully influence qPCR-based mtDNA copy number determination. Mitochondrion. 2009; 9:261–265.
Article27. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43:D447–D452.
Article28. Garcia G, Godot V, Humbert M. New chemokine targets for asthma therapy. Curr Allergy Asthma Rep. 2005; 5:155–160.
Article29. Cannistraci CV, Ogorevc J, Zorc M, Ravasi T, Dovc P, Kunej T. Pivotal role of the muscle-contraction pathway in cryptorchidism and evidence for genomic connections with cardiomyopathy pathways in RASopathies. BMC Med Genomics. 2013; 6:5.
Article30. Li H, Chiappinelli KB, Guzzetta AA, Easwaran H, Yen RW, Vatapalli R, Topper MJ, Luo J, Connolly RM, Azad NS, Stearns V, Pardoll DM, Davidson N, Jones PA, Slamon DJ, Baylin SB, Zahnow CA, Ahuja N. Immune regulation by low doses of the DNA methyltransferase inhibitor 5-azacitidine in common human epithelial cancers. Oncotarget. 2014; 5:587–598.
Article31. Forner F, Kumar C, Luber CA, Fromme T, Klingenspor M, Mann M. Proteome differences between brown and white fat mitochondria reveal specialized metabolic functions. Cell Metab. 2009; 10:324–335.
Article32. Falk MJ, Zhang Z, Rosenjack JR, Nissim I, Daikhin E, Nissim I, Sedensky MM, Yudkoff M, Morgan PG. Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Mol Genet Metab. 2008; 93:388–397.
Article33. Jaakkola MS, Ieromnimon A, Jaakkola JJ. Are atopy and specific IgE to mites and molds important for adult asthma? J Allergy Clin Immunol. 2006; 117:642–648.
Article34. Sebastiani S, Albanesi C, De PO, Puddu P, Cavani A, Girolomoni G. The role of chemokines in allergic contact dermatitis. Arch Dermatol Res. 2002; 293:552–559.
Article35. Bao L, Shi VY, Chan LS. IL-4 regulates chemokine CCL26 in keratinocytes through the Jak1, 2/Stat6 signal transduction pathway: Implication for atopic dermatitis. Mol Immunol. 2012; 50:91–97.
Article36. Danielewicz H. Hits and defeats of genome-wide association studies of atopy and asthma. J Appl Biomed. 2017; 15:161–168.
Article37. Grammatikos AP. The genetic and environmental basis of atopic diseases. Ann Med. 2008; 40:482–495.
Article38. Robbins SH, Walzer T, Dembélé D, Thibault C, Defays A, Bessou G, Xu H, Vivier E, Sellars M, Pierre P, Sharp FR, Chan S, Kastner P, Dalod M. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008; 9:R17.
Article39. Li H, Toh PZ, Tan JY, Zin MT, Lee CY, Li B, Leolukman M, Bao H, Kang L. Selected biomarkers revealed potential skin toxicity caused by certain copper compounds. Sci Rep. 2016; 6:37664.
Article40. Cakebread JA, Haitchi HM, Holloway JW, Powell RM, Keith T, Davies DE, Holgate ST. The role of ADAM33 in the pathogenesis of asthma. Springer Semin Immunopathol. 2004; 25:361–375.
Article41. Pinto LA, Steudemann L, Depner M, Klopp N, Illig T, Weiland SK, von Mutius E, Kabesch M. STAT1 gene variations, IgE regulation and atopy. Allergy. 2007; 62:1456–1461.
Article42. Johansen C, Rittig AH, Mose M, Bertelsen T, Weimar I, Nielsen J, Andersen T, Rasmussen TK, Deleuran B, Iversen L. STAT2 is involved in the pathogenesis of psoriasis by promoting CXCL11 and CCL5 production by keratinocytes. PLoS One. 2017; 12:e0176994.
Article43. Zaidi N, Swinnen JV, Smans K. ATP-citrate lyase: a key player in cancer metabolisms. Cancer Res. 2012; 72:3709–3714.44. Infantino V, Iacobazzi V, Palmieri F, Menga A. ATP-citrate lyase is essential for macrophage inflammatory response. Biochem Biophys Res Commun. 2013; 440:105–111.
Article45. Millward CA, Desantis D, Hsieh CW, Heaney JD, Pisano S, Olswang Y, Reshef L, Beidelschies M, Puchowicz M, Croniger CM. Phosphoenolpyruvate carboxykinase (Pck1) helps regulate the triglyceride/ fatty acid cycle and development of insulin resistance in mice. J Lipid Res. 2010; 51:1452–1463.46. Porter C, Herndon DN, Chondronikola M, Chao T, Annamalai P, Bhattarai N, Saraf MK, Capek KD, Reidy PT, Daquinag AC, Kolonin MG, Rasmussen BB, Borsheim E, Toliver-Kinsky T, Sidossis LS. Human and mouse brown adipose tissue mitochondria have comparable UCP1 function. Cell Metab. 2016; 24:246–255.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- A Novel Model for Human Atopic Dermatitis: Application of Repeated DNCB Patch in BALB/c Mice, in Comparison with NC/Nga Mice
- Developing an Atopic Dermatitis Model and the Effects of Actinidia Extract on Dermatitis in NC/Nga Mice
- Alteration of Innate Immune T and B Cells in the NC/Nga Mouse
- A Study on Tests of Skin Safety and Inhibition of Atopic Dermatitis Using a StoneTouch(R) Infrared Scanner in a Mouse Model
- Screening of Anti-Atopic Dermatitis Material by Using NC/Nga Mouse Whole Blood System