Ann Lab Med.
2017 Nov;37(6):505-510. 10.3343/alm.2017.37.6.505.
Utility of a Direct 16S rDNA PCR and Sequencing for Etiological Diagnosis of Infective Endocarditis
- Affiliations
-
- 1Department of Laboratory Medicine, Asan Medical Center and University of Ulsan College of Medicine, Seoul, Korea. sung@amc.seoul.kr
- 2Department of Internal Medicine, Asan Medical Center and University of Ulsan College of Medicine, Seoul, Korea.
- 3Department of Thoracic and Cardiovascular Surgery, Asan Medical Center and University of Ulsan College of Medicine, Seoul, Korea.
- KMID: 2425991
- DOI: http://doi.org/10.3343/alm.2017.37.6.505
Abstract
- BACKGROUND
Cases of infective endocarditis (IE) require prompt etiological diagnosis for effective treatment. Molecular methods can aid in rapid and reliable diagnosis of culture-negative IE cases. We evaluated the utility of 16S rDNA PCR and sequencing in determining the causative agents of IE in valve tissues, especially when specimens were obtained after initiation of antimicrobial therapy.
METHODS
We performed 16S rDNA PCR and sequencing in heart valve specimens and medical records review of 80 patients who underwent protocol-based cardiac surgery from 2013 to 2015. One patient did not meet the criteria for IE. Sixty-five (81.3%) and 14 pa-tients (17.5%) were diagnosed as having definite IE and possible IE, respectively. Blood and heart valve biopsy tissue were examined by using routine microbiological methods.
RESULTS
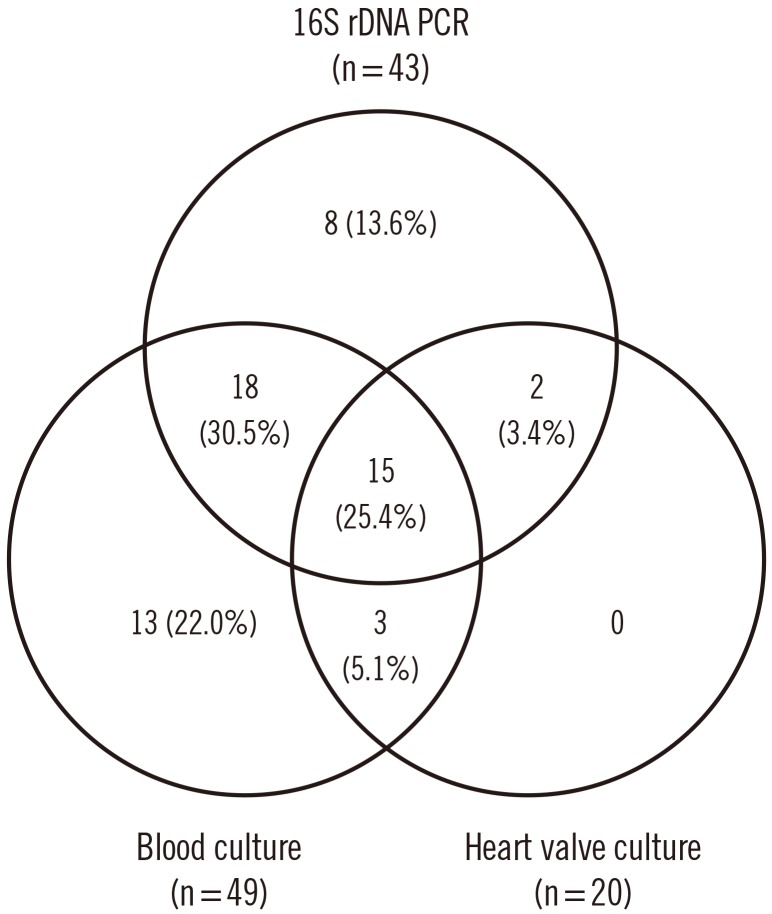
Blood cultures in our hospital were IE-positive for 33 patients (41.8%), whereas 49 patients (62.0%) showed positive blood cultures when initial blood cultures performed at the referring hospital were included. Eighteen (22.8%) and 40 patients (50.6%) were IE-positive in valve tissue cultures and 16S rDNA PCR, respectively. Bacteria in the Streptococcus mitis group (n=26) were the most common etiological agents of IE. Eight (10.1%) culture-negative specimens tested positive by 16S rDNA PCR. In five of eight PCR-positive and culture-negative cases, fastidious or anaerobic organisms were the cause of IE.
CONCLUSIONS
Direct 16S rDNA PCR and sequencing can be used as a supplementary method to conventional blood and biopsy culture testing, especially in culture-negative IE cases that are negative for IE by culture.
Keyword
MeSH Terms
Figure
Reference
-
1. Wouters BJ, Sanders MA, Lugthart S, Geertsma-Kleinekoort WM, van Drunen E, Beverloo HB, et al. Segmental uniparental disomy as a recurrent mechanism for homozygous CEBPA mutations in acute myeloid leukemia. Leukemia. 2007; 21:2382–2384. PMID: 17554374.2. Lamas CC, Eykyn SJ. Blood culture negative endocarditis: analysis of 63 cases presenting over 25 years. Heart. 2003; 89:258–262. PMID: 12591823.3. Gould FK, Denning DW, Elliott TS, Foweraker J, Perry JD, Prendergast BD, et al. Guidelines for the diagnosis and antibiotic treatment of endocarditis in adults: a report of the Working Party of the British Society for Antimicrobial Chemotherapy. J Antimicrob Chemother. 2012; 67:269–289. PMID: 22086858.4. Nikkari S, Merilahti-Palo R, Saario R, Söderström KO, Granfors K, Skurnik M, et al. Yersinia-triggered reactive arthritis. Use of polymerase chain reaction and immunocytochemical staining in the detection of bacterial components from synovial specimens. Arthritis Rheum. 1992; 35:682–687. PMID: 1599522.5. Kotilainen P, Jalava J, Meurman O, Lehtonen OP, Rintala E, Seppälä OP, et al. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J Clin Microbiol. 1998; 36:2205–2209. PMID: 9665992.6. Rantakokko-Jalava K, Nikkari S, Jalava J, Eerola E, Skurnik M, Meurman O, et al. Direct amplification of rRNA genes in diagnosis of bacterial infections. J Clin Microbiol. 2000; 38:32–39. PMID: 10618059.7. Greub G, Lepidi H, Rovery C, Casalta JP, Habib G, Collard F, et al. Diagnosis of infectious endocarditis in patients undergoing valve surgery. Am J Med. 2005; 118:230–238. PMID: 15745720.8. Rovery C, Greub G, Lepidi H, Casalta JP, Habib G, Collart F, et al. PCR detection of bacteria on cardiac valves of patients with treated bacterial endocarditis. J Clin Microbiol. 2005; 43:163–167. PMID: 15634966.9. Houpikian P, Raoult D. Blood culture-negative endocarditis in a reference center: etiologic diagnosis of 348 cases. Medicine (Baltimore). 2005; 84:162–173. PMID: 15879906.10. Li JS, Sexton DJ, Mick N, Nettles R, Fowler VG Jr, Ryan T, et al. Proposed modifications to the Duke criteria for the diagnosis of infective endocarditis. Clin Infect Dis. 2000; 30:633–638. PMID: 10770721.11. Durack DT, Lukes AS, Bright DK. New criteria for diagnosis of infective endocarditis: utilization of specific echocardiographic findings. Duke Endocarditis Service. Am J Med. 1994; 96:200–209. PMID: 8154507.12. Relman DA. Universal bacterial 16S rDNA amplification and sequencing. Diagnostic molecular microbiology: principles and applications. Washington DC: American Society for Microbiology;1993. p. 489–495.13. Choi SH, Sung H, Kim SH, Lee SO, Lee SH, Kim YS, et al. Usefulness of a direct 16S rRNA gene PCR assay of percutaneous biopsies or aspirates for etiological diagnosis of vertebral osteomyelitis. Diagn Microbiol Infect Dis. 2014; 78:75–78. PMID: 24231384.14. CLSI. Interpretive criteria for identification of bacteria and fungi by dna target sequencing; Approved guideline MM18-A. Wayne, PA: CLSI;2008.15. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012; 62:716–721. PMID: 22140171.16. Park KS, Ki CS, Kang CI, Kim YJ, Chung DR, Peck KR, et al. Evaluation of the GenBank, EzTaxon, and BIBI services for molecular identification of clinical blood culture isolates that were unidentifiable or misidentified by conventional methods. J Clin Microbiol. 2012; 50:1792–1795. PMID: 22403421.17. Pant S, Patel NJ, Deshmukh A, Golwala H, Patel N, Badheka A, et al. Trends in infective endocarditis incidence, microbiology, and valve replacement in the United States from 2000 to 2011. J Am Coll Cardiol. 2015; 65:2070–2076. PMID: 25975469.18. Miller RJ, Chow B, Pillai D, Church D. Development and evaluation of a novel fast broad-range 16S ribosomal DNA PCR and sequencing assay for diagnosis of bacterial infective endocarditis: multi-year experience in a large Canadian healthcare zone and a literature review. BMC Infect Dis. 2016; 16:146. PMID: 27066823.19. Kotilainen P, Heiro M, Jalava J, Rantakokko V, Nikoskelainen J, Nikkari S, et al. Aetiological diagnosis of infective endocarditis by direct amplification of rRNA genes from surgically removed valve tissue. An 11-year experience in a Finnish teaching hospital. Ann Med. 2006; 38:263–273. PMID: 16754257.20. Fournier PE, Watt G, et al. Blood culture-negative endocarditis. In : Habib G, editor. Infective endocarditis: epidemiology, diagnosis, imaging, therapy, and prevention. 1st ed. Switzerland: Springer International Publishing AG;2016. p. 245–258.21. Horstkotte D, Follath F, Gutschik E, Lengyel M, Oto A, Pavie A, et al. Guidelines on prevention, diagnosis and treatment of infective endocarditis executive summary; the task force on infective endocarditis of the European society of cardiology. Eur Heart J. 2004; 25:267–276. PMID: 14972429.22. Task Force sull'ndocardite Infettiva della Societá Europea di Cardiologia. Guidelines on prevention, diagnosis and treatment of infective endocarditis. Ital Heart J Suppl. 2004; 5:548–590. PMID: 15490689.23. Morris AJ, Wilson SJ, Marx CE, Wilson ML, Mirrett S, Reller LB. Clinical impact of bacteria and fungi recovered only from broth cultures. J Clin Microbiol. 1995; 33:161–165. PMID: 7699035.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Haemophilus parainfluenzae Infective Endocarditis Diagnosed by Direct 16S rRNA Sequencing of Vegetation
- Identification of Gemella species by 16S ribosomal RNA gene sequencing from two patients with infective endocarditis
- A Case of Streptococcus gallolyticus subsp. gallolyticus Infective Endocarditis with Colon Cancer: Identification by 16S Ribosomal DNA Sequencing
- Haemophilus parainfluenzae Infective Endocarditis Confirmed by 16S rRNA Sequence Analysis from Culture Negative Tissue
- A Case of Infective Endocarditis Caused by Kytococcus schroeteri