Korean Circ J.
2014 Jul;44(4):255-263. 10.4070/kcj.2014.44.4.255.
miR-18a-5p MicroRNA Increases Vascular Smooth Muscle Cell Differentiation by Downregulating Syndecan4
- Affiliations
-
- 1Heart Research Center of Chonnam National University Hospital, Gwangju, Korea. myungho@chollian.net
- 2Division of Cardiovascular and Rare Diseases, Center for Biomedical Sciences, Korea National Institute of Health, Cheongju, Korea.
- 3Department of Pharmacology and Medical Research Center for Gene Regulation, Chonnam National University Medical School, Gwangju, Korea.
- KMID: 2223887
- DOI: http://doi.org/10.4070/kcj.2014.44.4.255
Abstract
- BACKGROUND AND OBJECTIVES
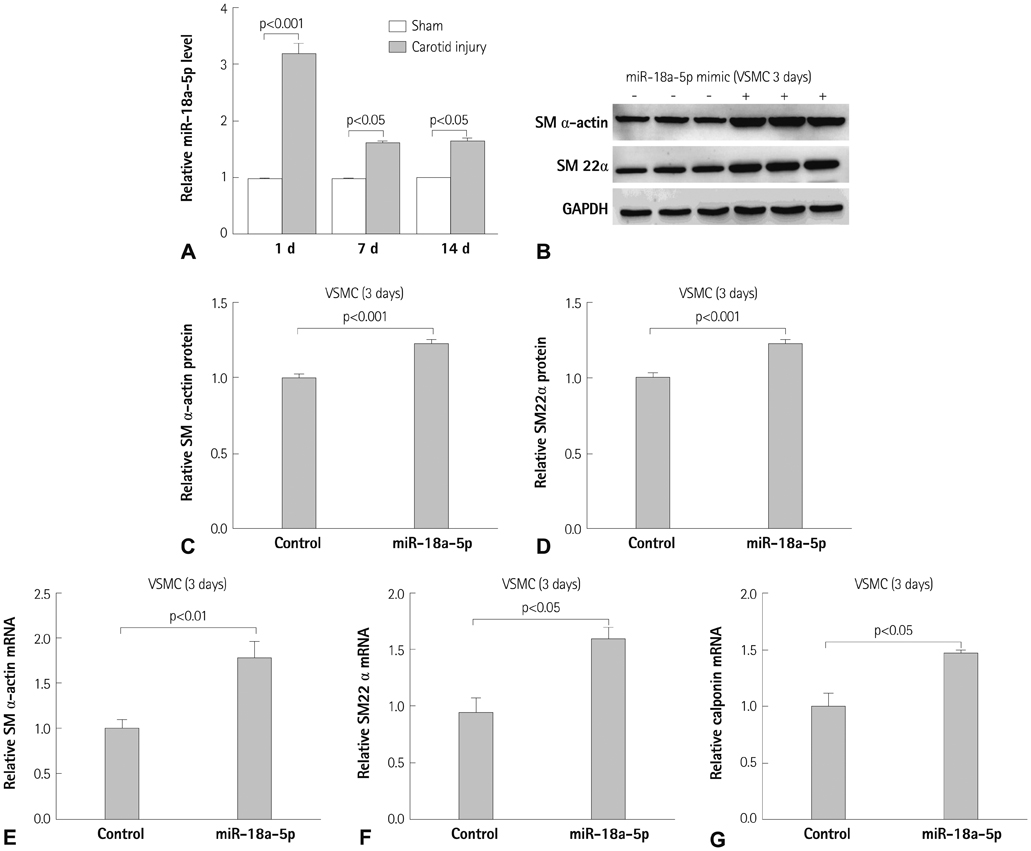
Differentiation and de-differentiation of vascular smooth muscle cells (VSMCs) are important events in atherosclerosis and restenosis after angioplasty. MicroRNAs are considered a key regulator in cellular processes such as differentiation, proliferation, and apoptosis. Here, we report the role of new miR-18a-5p microRNA and its downstream target genes in VSMCs and in a carotid balloon injury model.
MATERIALS AND METHODS
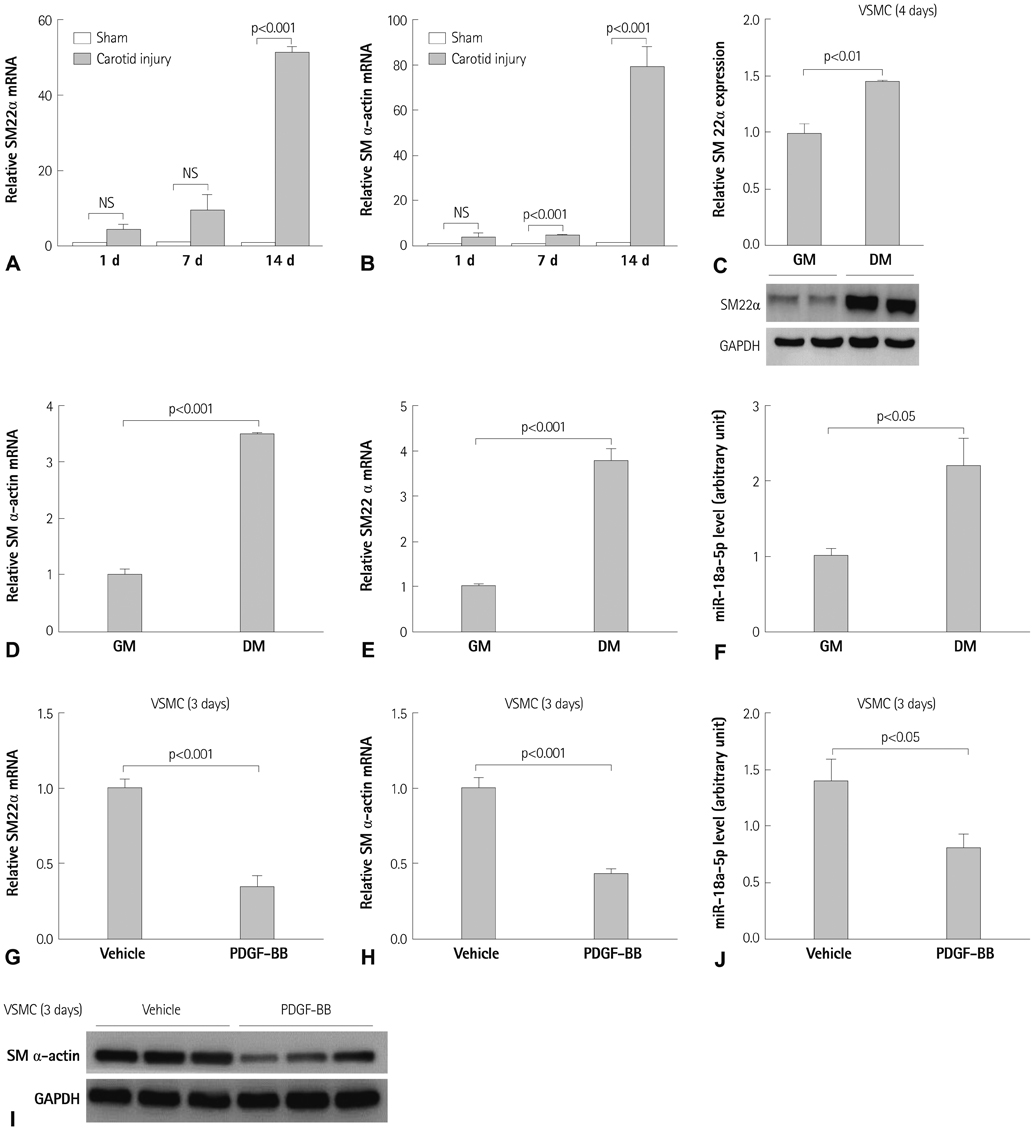
Expression of miR-18a-5p and its candidate genes was examined in VSMCs and in a carotid artery injury model by quantitative reverse-transcription polymerase chain reaction (qRT-PCR) and microRNA microarray analysis. VSMC differentiation marker genes including smooth muscle (SM) alpha-actin and SM22alpha were determined by Western blot, qRT-PCR, and a SM22alpha promoter study. Gene overexpression or knockdown was performed in VSMCs.
RESULTS
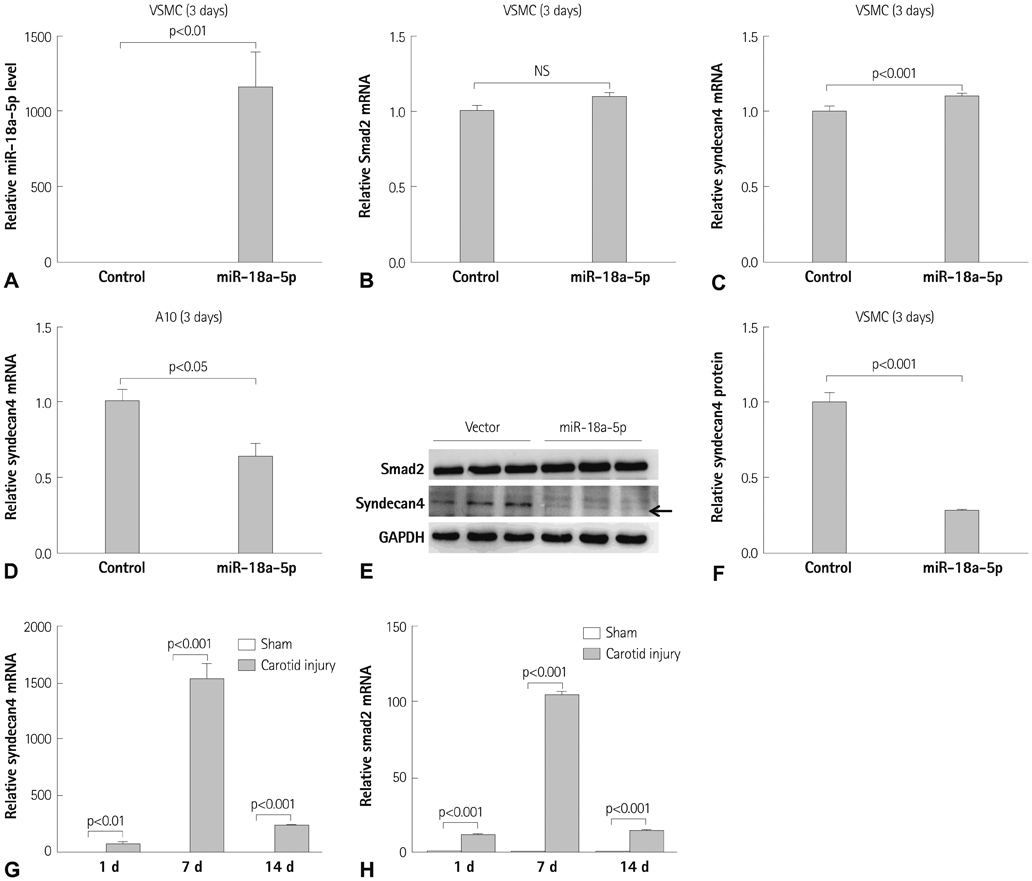
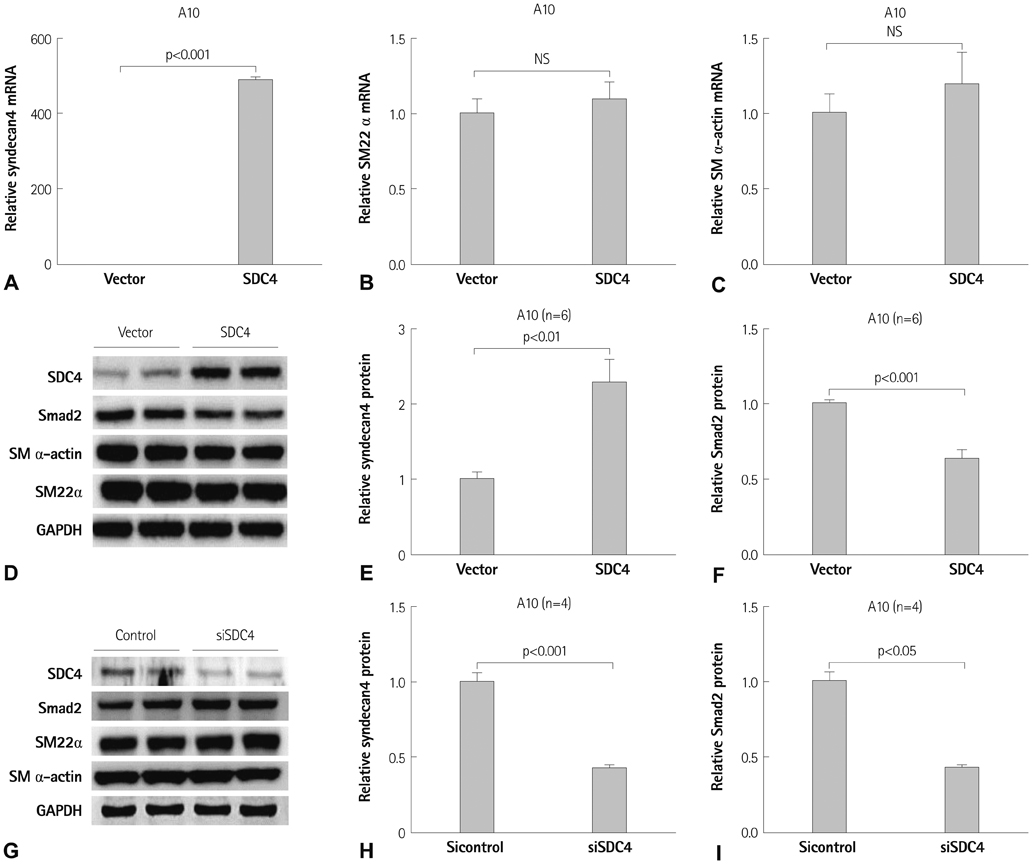
miR-18a-5p was upregulated in the rat carotid artery at the early time after balloon injury. Transfection of the miR-18a-5p mimic promoted the VSMC differentiation markers SM alpha-actin and SM22alpha. In addition, miR-18a-5p expression was induced in differentiated VSMCs, whereas it decreased in de-differentiated VSMCs. We identified syndecan4 as a downstream target of miR-18-5p in VSMCs. Overexpression of syndecan4 decreased Smad2 expression, whereas knockdown of syndecan4 increased Smad2 expression in VSMCs. Finally, we showed that Smad2 induced the expression of VSMC differentiation marker genes in VSMCs.
CONCLUSION
These results indicate that miR-18a-5p is involved in VSMC differentiation by targeting syndecan4.
MeSH Terms
-
Actins
Angioplasty
Animals
Antigens, Differentiation
Apoptosis
Atherosclerosis
Blotting, Western
Carotid Arteries
Carotid Artery Injuries
Cell Differentiation*
Microarray Analysis
MicroRNAs*
Muscle, Smooth
Muscle, Smooth, Vascular*
Polymerase Chain Reaction
Rats
Smad2 Protein
Syndecan-4*
Transfection
Actins
Antigens, Differentiation
MicroRNAs
Smad2 Protein
Syndecan-4
Figure
Reference
-
1. Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008; 28:812–819.2. Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004; 84:767–801.3. Ambros V. The functions of animal microRNAs. Nature. 2004; 431:350–355.4. He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat Rev Genet. 2004; 5:522–531.5. Cordes KR, Sheehy NT, White MP, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009; 460:705–710.6. Cheng Y, Liu X, Yang J, et al. MicroRNA-145, a novel smooth muscle cell phenotypic marker and modulator, controls vascular neointimal lesion formation. Circ Res. 2009; 105:158–166.7. Li P, Zhu N, Yi B, et al. MicroRNA-663 regulates human vascular smooth muscle cell phenotypic switch and vascular neointimal formation. Circ Res. 2013; 113:1117–1127.8. Leeper NJ, Raiesdana A, Kojima Y, et al. MicroRNA-26a is a novel regulator of vascular smooth muscle cell function. J Cell Physiol. 2011; 226:1035–1043.9. Torella D, Iaconetti C, Catalucci D, et al. MicroRNA-133 controls vascular smooth muscle cell phenotypic switch in vitro and vascular remodeling in vivo. Circ Res. 2011; 109:880–893.10. Davis BN, Hilyard AC, Nguyen PH, Lagna G, Hata A. Induction of microRNA-221 by platelet-derived growth factor signaling is critical for modulation of vascular smooth muscle phenotype. J Biol Chem. 2009; 284:3728–3738.11. Choe N, Kwon JS, Kim JR, et al. The microRNA miR-132 targets Lrrfip1 to block vascular smooth muscle cell proliferation and neointimal hyperplasia. Atherosclerosis. 2013; 229:348–355.12. Friedman LM, Dror AA, Mor E, et al. MicroRNAs are essential for development and function of inner ear hair cells in vertebrates. Proc Natl Acad Sci U S A. 2009; 106:7915–7920.13. Hayashita Y, Osada H, Tatematsu Y, et al. A polycistronic microRNA cluster, miR-17-92, is overexpressed in human lung cancers and enhances cell proliferation. Cancer Res. 2005; 65:9628–9632.14. Ohgawara T, Kubota S, Kawaki H, et al. Regulation of chondrocytic phenotype by micro RNA 18a: involvement of Ccn2/Ctgf as a major target gene. FEBS Lett. 2009; 583:1006–1010.15. Kee HJ, Kwon JS, Shin S, Ahn Y, Jeong MH, Kook H. Trichostatin A prevents neointimal hyperplasia via activation of Krüppel like factor 4. Vascul Pharmacol. 2011; 55:127–134.16. Kwon JS, Joung H, Kim YS, et al. Sulforaphane inhibits restenosis by suppressing inflammation and the proliferation of vascular smooth muscle cells. Atherosclerosis. 2012; 225:41–49.17. Ikesue M, Matsui Y, Ohta D, et al. Syndecan-4 deficiency limits neointimal formation after vascular injury by regulating vascular smooth muscle cell proliferation and vascular progenitor cell mobilization. Arterioscler Thromb Vasc Biol. 2011; 31:1066–1074.18. Matsui Y, Ikesue M, Danzaki K, et al. Syndecan-4 prevents cardiac rupture and dysfunction after myocardial infarction. Circ Res. 2011; 108:1328–1339.19. Herum KM, Lunde IG, Skrbic B, et al. Syndecan-4 signaling via NFAT regulates extracellular matrix production and cardiac myofibroblast differentiation in response to mechanical stress. J Mol Cell Cardiol. 2013; 54:73–81.20. Xie WB, Li Z, Shi N, et al. Smad2 and myocardin-related transcription factor B cooperatively regulate vascular smooth muscle differentiation from neural crest cells. Circ Res. 2013; 113:e76–e86.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- MicroRNA-139-5p Regulates Fibrotic Potentials via Modulation of Collagen Type 1 and Phosphorylated p38 MAPK in Uterine Leiomyoma
- Lactate promotes vascular smooth muscle cell switch to a synthetic phenotype by inhibiting miR-23b expression
- Differentially expressed mRNAs and their upstream miR-491-5p in patients with coronary atherosclerosis as well as the function of miR-491-5p in vascular smooth muscle cells
- MicroRNA-181a-5p Curbs Osteogenic Differentiation and Bone Formation Partially Through Impairing Runx1-Dependent Inhibition of AIF-1 Transcription
- Expression of miR-221 and miR-18a in patients with hepatocellular carcinoma and its clinical significance