Ann Dermatol.
2020 Apr;32(2):130-140. 10.5021/ad.2020.32.2.130.
Identification of Molecular Signatures in Mild Intrinsic Atopic Dermatitis by Bioinformatics Analysis
- Affiliations
-
- 1Department of Dermatology, Fu Dan University, Huashan Hospital, Shanghai, China. guchaoy99@163.com
- KMID: 2471339
- DOI: http://doi.org/10.5021/ad.2020.32.2.130
Abstract
- BACKGROUND
Atopic dermatitis (AD) is recognized as a common inflammatory skin disease and frequently occurred in Asian and Black individuals.
OBJECTIVE
Since the limitation of dataset associated with human severe AD, this study aimed to screen potential novel biomarkers involved in mild AD.
METHODS
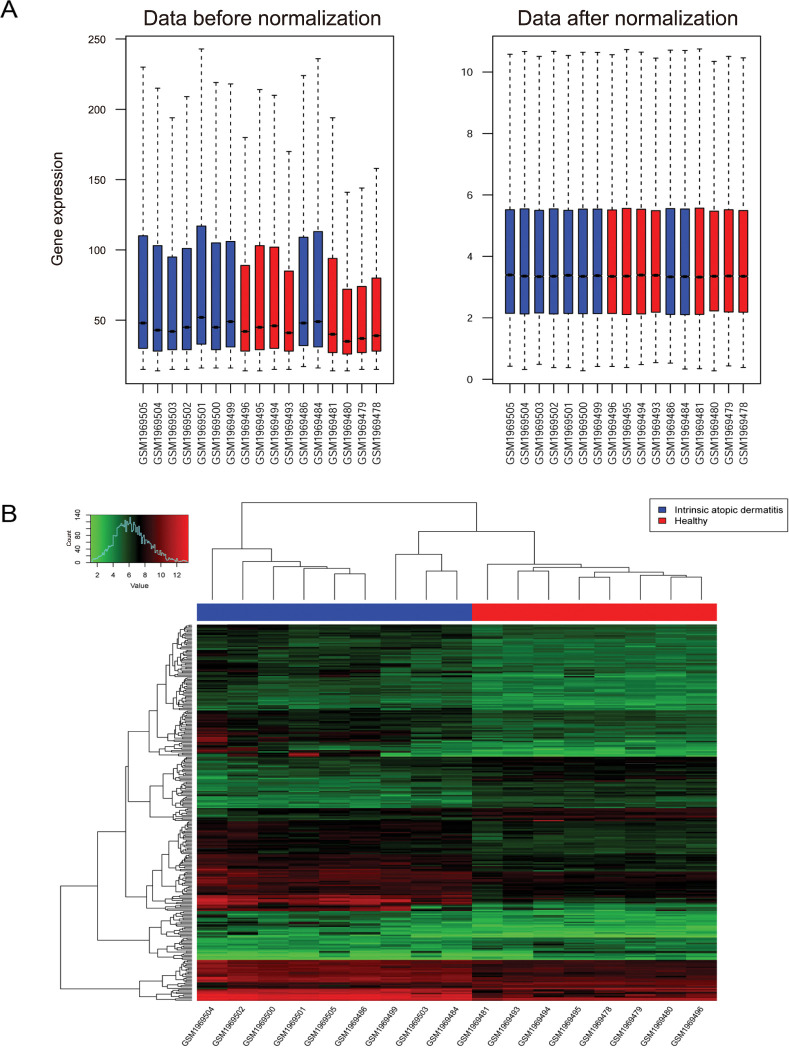
Expression profile data (GSE75890) were obtained from the database of Gene Expression Omnibus. Using limma package, the differentially expressed genes (DEGs) between samples from AD and healthy control were selected. Furthermore, function analysis was conducted. Meanwhile, the protein-protein interaction (PPI) network and transcription factor (TF)-miRNA-target regulatory network were constructed. And quantitative real-time polymerase chain reaction (qRT-PCR) was used to validate the expressions patterns of key genes.
RESULTS
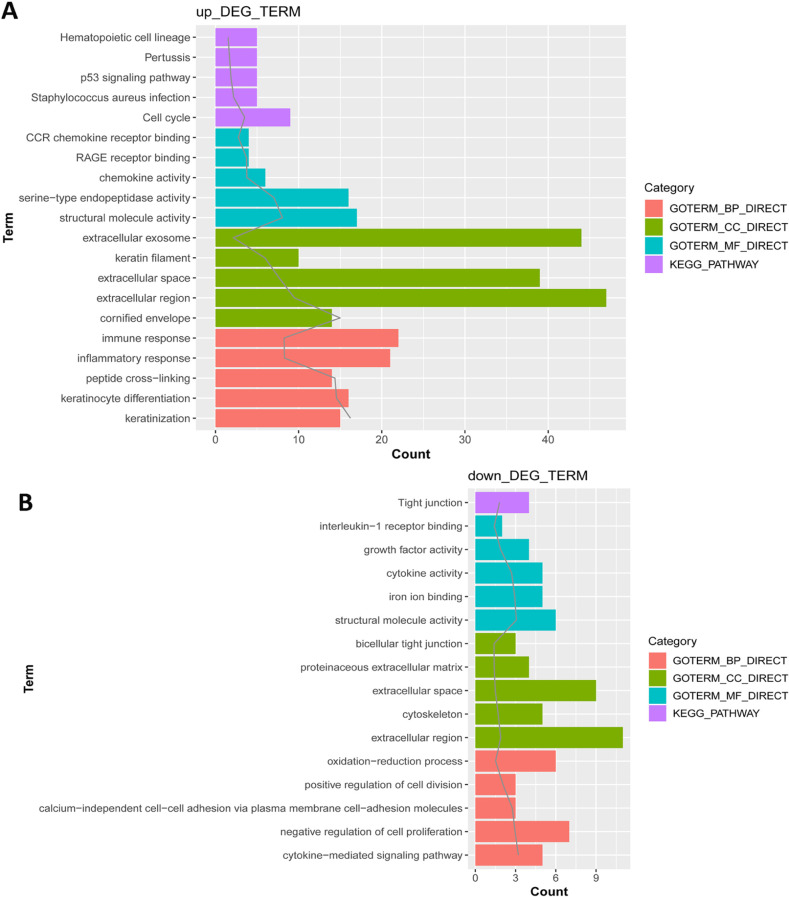
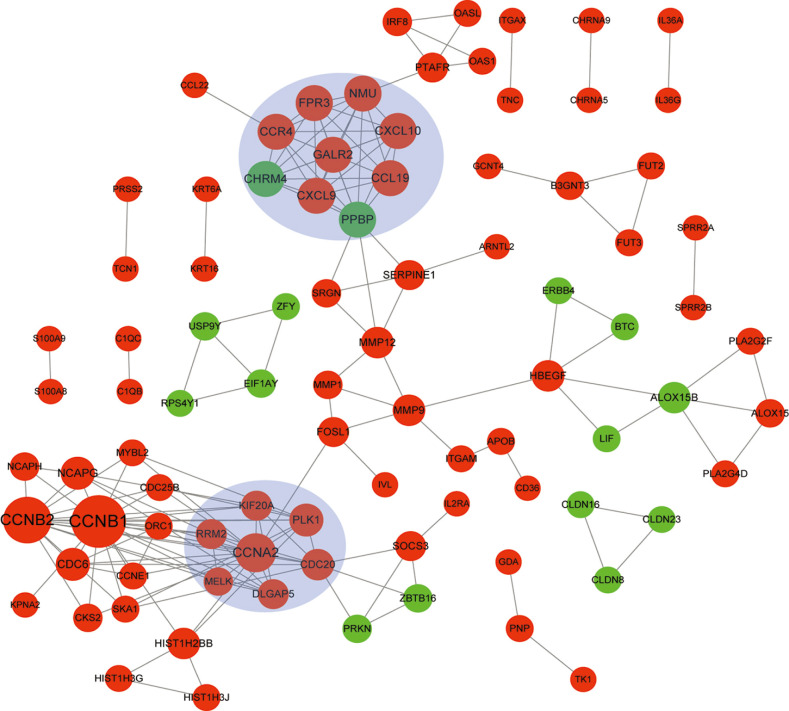
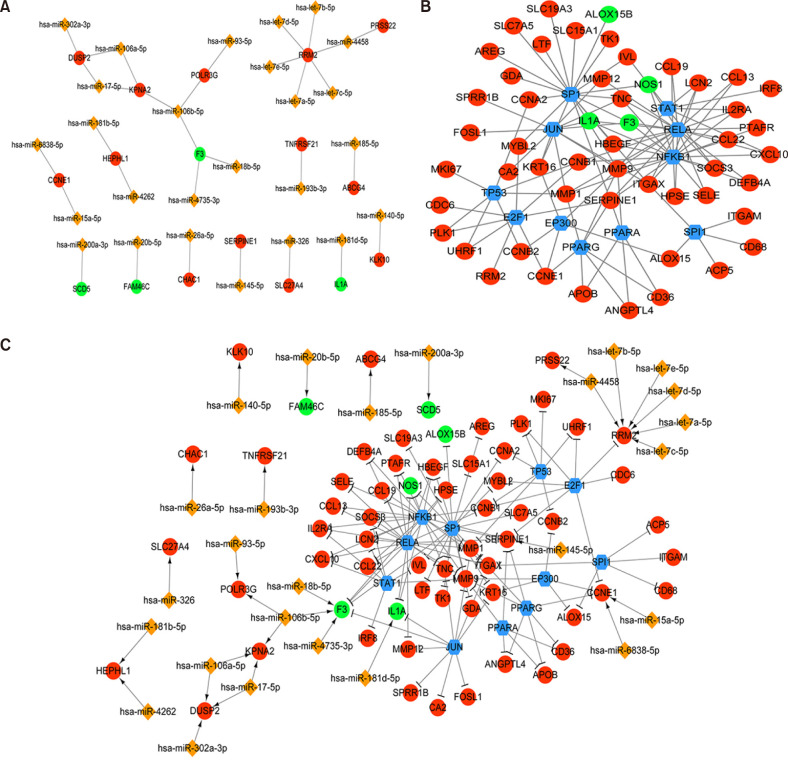
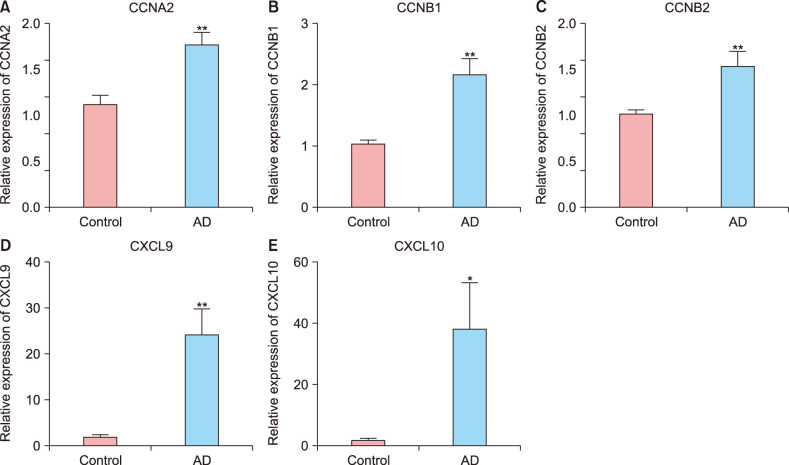
In total, 285 DEGs including 214 upregulated and 71 downregulated genes were identified between samples from two groups. The upregulated DEGs were mainly involved in nine pathways, such as hematopoietic cell lineage, pertussis, p53 signaling pathway, staphylococcus aureus infection, and cell cycle, while tight junction was the only pathway enriched by the downregulated DEGs. Cyclin B (CCNB)1, CCNB2, cyclin A (CCNA)2, C-X-C motif chemokine ligand (CXCL)10, and CXCL9 were key nodes in PPI network. The TF-miRNA-target gene regulatory network focused on miRNAs such as miR-106b, miR-106a, and miR-17, TFs such as nuclear factor kappa B subunit 1, RELA proto-oncogene, Sp1 transcription factor, and genes such as matrix metallopeptidase 9, peroxisome proliferator activated receptor gamma , and serpin family E member 1. Moreover, the upregulation of these genes, including CCNB1, CCNB2, CCNA2, CXCL10, and CXCL9 were confirmed by qRT-PCR.
CONCLUSION
CCNB1, CCNB2, CCNA2, and CXCL9 might be novel markers of mild AD. miR-106b and miR-17 may involve in regulation of immune response in AD patients.
Keyword
MeSH Terms
-
Asian Continental Ancestry Group
Biomarkers
Cell Cycle
Cell Lineage
Computational Biology*
Cyclin A
Cyclin B
Dataset
Dermatitis
Dermatitis, Atopic*
Gene Expression
Gene Regulatory Networks
Humans
MicroRNAs
NF-kappa B
PPAR gamma
Proto-Oncogenes
Real-Time Polymerase Chain Reaction
Skin Diseases
Sp1 Transcription Factor
Staphylococcus aureus
Tight Junctions
Transcription Factors
Up-Regulation
Whooping Cough
Biomarkers
Cyclin A
Cyclin B
MicroRNAs
NF-kappa B
PPAR gamma
Sp1 Transcription Factor
Transcription Factors
Figure
Reference
-
1. Hua T, Silverberg JI. Atopic dermatitis in US adults: epidemiology, association with marital status, and atopy. Ann Allergy Asthma Immunol. 2018; 121:622–624. PMID: 30036584.2. Sacotte R, Silverberg JI. Epidemiology of adult atopic dermatitis. Clin Dermatol. 2018; 36:595–605. PMID: 30217272.
Article3. Kaufman BP, Guttman-Yassky E, Alexis AF. Atopic dermatitis in diverse racial and ethnic groups-Variations in epidemiology, genetics, clinical presentation and treatment. Exp Dermatol. 2018; 27:340–357. PMID: 29457272.
Article4. Rahman S, Collins M, Williams CM, Ma HL. The pathology and immunology of atopic dermatitis. Inflamm Allergy Drug Targets. 2011; 10:486–496. PMID: 21864272.
Article5. Pavel AB, Song T, Kim HJ, Del Duca E, Krueger JG, Dubin C, et al. Oral Janus kinase/SYK inhibition (ASN002) suppresses inflammation and improves epidermal barrier markers in patients with atopic dermatitis. J Allergy Clin Immunol. 2019; 144:1011–1024. PMID: 31356921.
Article6. Guttman-Yassky E, Bissonnette R, Ungar B, Suárez-Fariñas M, Ardeleanu M, Esaki H, et al. Dupilumab progressively improves systemic and cutaneous abnormalities in patients with atopic dermatitis. J Allergy Clin Immunol. 2019; 143:155–172. PMID: 30194992.
Article7. Beck LA, Thaçi D, Hamilton JD, Graham NM, Bieber T, Rocklin R, et al. Dupilumab treatment in adults with moderate-to-severe atopic dermatitis. N Engl J Med. 2014; 371:130–139. PMID: 25006719.
Article8. Lee HJ, Lee NR, Jung M, Kim DH, Choi EH. Atopic march from atopic dermatitis to asthma-like lesions in NC/Nga mice is accelerated or aggravated by neutralization of stratum corneum but partially inhibited by acidification. J Invest Dermatol. 2015; 135:3025–3033. PMID: 26399697.
Article9. Sonkoly E, Janson P, Majuri ML, Savinko T, Fyhrquist N, Eidsmo L, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010; 126:581589.e1–589.e20. PMID: 20673989.
Article10. Martel BC, Litman T, Hald A, Norsgaard H, Lovato P, Dyring-Andersen B, et al. Distinct molecular signatures of mild extrinsic and intrinsic atopic dermatitis. Exp Dermatol. 2016; 25:453–459. PMID: 26841714.
Article11. Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010; 26:2363–2367. PMID: 20688976.
Article12. Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003; 19:185–193. PMID: 12538238.
Article13. Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003; 4:249–264. PMID: 12925520.
Article14. Smyth GK. Limma: linear models for microarray data. In : Gentleman R, editor. Bioinformatics and computational biology solutions using R and bioconductor. New York: Springer-Verlag;2005. p. 397–420.15. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57. PMID: 19131956.
Article16. Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000; 28:27–30. PMID: 10592173.
Article17. Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015; 43(Database issue):D447–D452. PMID: 25352553.
Article18. Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003; 13:2498–2504. PMID: 14597658.
Article19. Bandettini WP, Kellman P, Mancini C, Booker OJ, Vasu S, Leung SW, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: a clinical validation study. J Cardiovasc Magn Reson. 2012; 14:83. PMID: 23199362.
Article20. Dweep H, Sticht C, Pandey P, Gretz N. miRWalk--database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011; 44:839–847. PMID: 21605702.21. Han H, Cho JW, Lee S, Yun A, Kim H, Bae D, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018; 46:D380–D386. PMID: 29087512.
Article22. Boguniewicz M, Leung DY. Atopic dermatitis: a disease of altered skin barrier and immune dysregulation. Immunol Rev. 2011; 242:233–246. PMID: 21682749.
Article23. Akdis M, Trautmann A, Klunker S, Daigle I, Kucuksezer UC, Deglmann W, et al. T helper (Th) 2 predominance in atopic diseases is due to preferential apoptosis of circulating memory/effector Th1 cells. FASEB J. 2003; 17:1026–1035. PMID: 12773485.
Article24. Grewe M, Bruijnzeel-Koomen CA, Schöpf E, Thepen T, Langeveld-Wildschut AG, Ruzicka T, et al. A role for Th1 and Th2 cells in the immunopathogenesis of atopic dermatitis. Immunol Today. 1998; 19:359–361. PMID: 9709503.
Article25. Shimada Y, Takehara K, Sato S. Both Th2 and Th1 chemokines (TARC/CCL17, MDC/CCL22, and Mig/CXCL9) are elevated in sera from patients with atopic dermatitis. J Dermatol Sci. 2004; 34:201–208. PMID: 15113590.
Article26. Abramo F, Campora L, Albanese F, della Valle MF, Cristino L, Petrosino S, et al. Increased levels of palmitoylethanolamide and other bioactive lipid mediators and enhanced local mast cell proliferation in canine atopic dermatitis. BMC Vet Res. 2014; 10:21. PMID: 24423192.
Article27. Meng K, Xu W, Miura T, Suzuki S, Chiyotanda M, Tanaka S, et al. The effects of vitamin K1 and vitamin K2 on the proliferation, cytokine production and regulatory T-cell frequency in peripheral blood mononuclear cells of paediatric atopic dermatitis patients. Exp Dermatol. 2018; 27:1058–1060. PMID: 29697859.
Article28. Rożalski M, Rudnicka L, Samochocki Z. MiRNA in atopic dermatitis. Postepy Dermatol Alergol. 2016; 33:157–162. PMID: 27512348.
Article29. Lv Y, Qi R, Xu J, Di Z, Zheng H, Huo W, et al. Profiling of serum and urinary microRNAs in children with atopic dermatitis. PLoS One. 2014; 9:e115448. PMID: 25531302.
Article30. Björkqvist J, de Maat S, Lewandrowski U, Di Gennaro A, Oschatz C, Schönig K, et al. Defective glycosylation of coagulation factor XII underlies hereditary angioedema type III. J Clin Invest. 2015; 125:3132–3146. PMID: 26193639.
Article31. Nguyen GN, Yamagata Y, Shigematsu Y, Watanabe M, Miyazaki Y, Doi K, et al. Duplication and loss of function of genes encoding RNA polymerase III subunit C4 causes hybrid incompatibility in rice. G3 (Bethesda). 2017; 7:2565–2575. PMID: 28592558.
Article32. Huang L, Zhou Y, Cao XP, Lin JX, Zhang L, Huang ST, et al. KPNA2 promotes migration and invasion in epithelial ovarian cancer cells by inducing epithelial-mesenchymal transition via Akt/GSK-3β/Snail activation. J Cancer. 2018; 9:157–165. PMID: 29290781.
Article33. Yamada K, Miyamoto Y, Tsujii A, Moriyama T, Ikuno Y, Shiromizu T, et al. Cell surface localization of importin α 1/KPNA2 affects cancer cell proliferation by regulating FGF1 signalling. Sci Rep. 2016; 6:21410. PMID: 26887791.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Age-related Changes in the Frequency of Intrinsic and Extrinsic Atopic Dermatitis: A Single-center Retrospective Study
- Measurement of Atopic Dermatitis Disability
- Serum IgE Level in Patients of Atopic Dermatitis and Atopic Dermatitis with Molluscum Contagiosum
- A Study on the Relationship of the Severity of Atopic Dermatitis, Serum IgE and IFN-gamma
- Nipple Involvement in Atopic Dermatitis: Report of 3 cases