Allergy Asthma Immunol Res.
2020 Mar;12(2):322-337. 10.4168/aair.2020.12.2.322.
Imbalance of Gut Streptococcus, Clostridium, and Akkermansia Determines the Natural Course of Atopic Dermatitis in Infant
- Affiliations
-
- 1Asan Institute for Life Sciences, Asan Medical Center, Seoul, Korea.
- 2Department of Pediatrics, Childhood Asthma Atopy Center, Environmental Health Center, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea. sjhong@amc.seoul.kr
- 3Environmental Health Center, Asan Medical Center, Seoul, Korea.
- 4Department of Life Science, Multidisciplinary Genome Institute, Hallym University, Chuncheon, Korea.
- 5Department of Pediatrics, Pusan National University Yangsan Hospital, Yangsan, Korea.
- 6Department of Pediatrics, Mediplex Sejong Hospital, Incheon, Korea.
- 7Department of Pediatrics, International St. Mary's Hospital, Catholic Kwandong University College of Medicine, Incheon, Korea.
- 8Department of Pediatrics, Chonnam National University Hospital, Chonnam National University Medical School, Gwangju, Korea.
- 9Department of Pediatrics, Hallym University Sacred Heart Hospital, Hallym University College of Medicine, Anyang, Korea.
- 10Department of Pediatrics, Dankuk University Hospital, Cheonan, Korea.
- 11Department of Pediatrics, Inje University Sanggye Paik Hospital, Seoul, Korea.
- 12Department of Pediatrics, Seoul National University College of Medicine, Seoul, Korea.
- 13Department of Pediatrics, CHA Gangnam Medical Center, CHA University School of Medicine, Seoul, Korea.
- 14Department of Pediatrics, Yonsei University of Medicine, Seoul, Korea.
- 15Department of Pediatrics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- KMID: 2468465
- DOI: http://doi.org/10.4168/aair.2020.12.2.322
Abstract
- PURPOSE
The roles of gut microbiota on the natural course of atopic dermatitis (AD) are not yet fully understood. We investigated whether the composition and function of gut microbiota and short-chain fatty acids (SCFAs) at 6 months of age could affect the natural course of AD up to 24 months in early childhood.
METHODS
Fecal samples from 132 infants were analyzed using pyrosequencing, including 84 healthy controls, 22 transient AD and 26 persistent AD subjects from the Cohort for Childhood Origin of Asthma and Allergic Diseases (COCOA) birth cohort. The functional profile of the gut microbiome was analyzed by whole-metagenome sequencing. SCFAs were measured using gas chromatography-mass spectrometry.
RESULTS
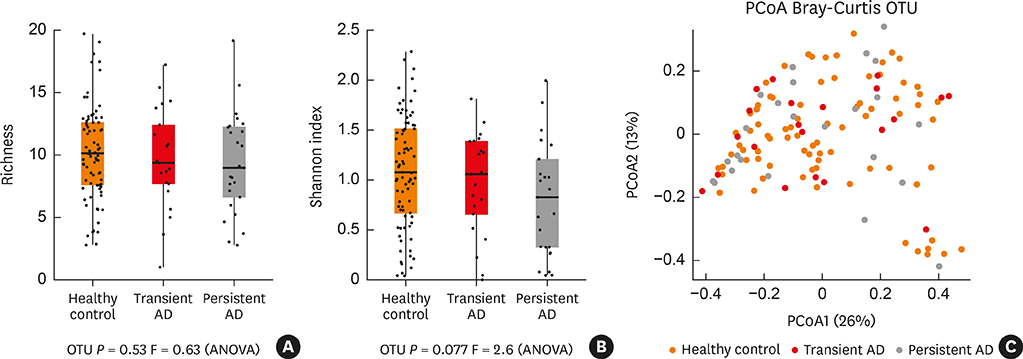
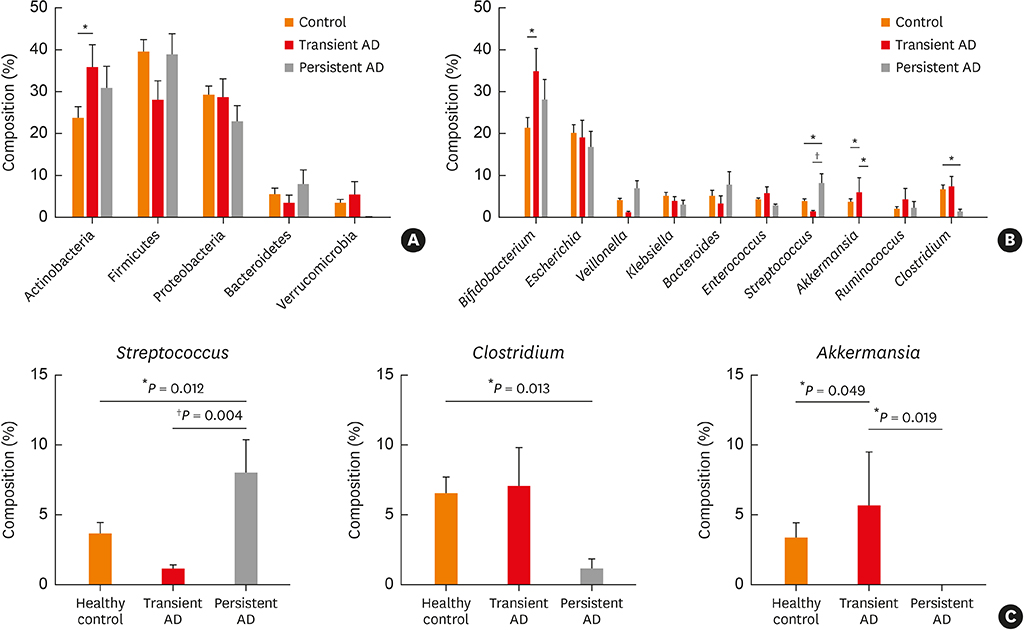
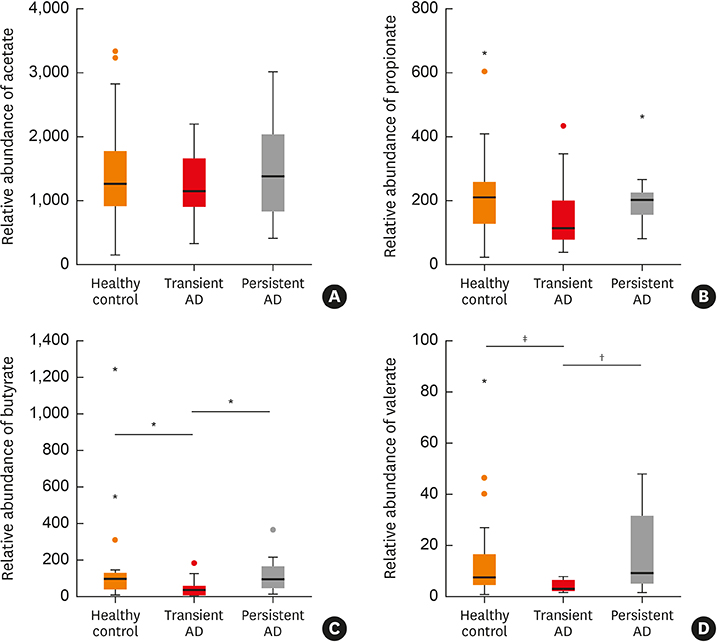
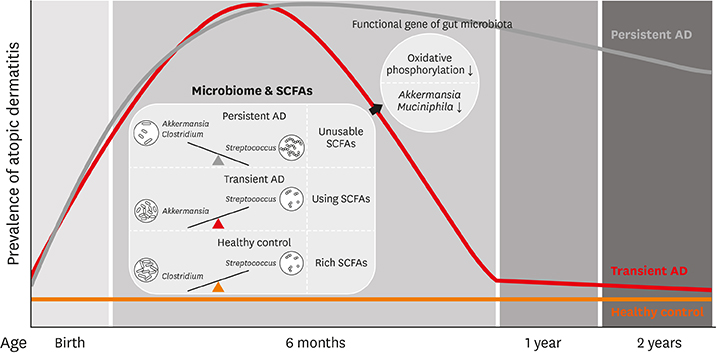
Low levels of Streptococcus and high amounts of Akkermansia were evident in transient AD cases, and low Clostridium, Akkermansia and high Streptococcus were found in children with persistent AD. The relative abundance of Streptococcus positively correlated with scoring of AD (SCORAD) score, whereas that of Clostridium negatively correlated with SCORAD score. The persistent AD group showed decreased gut microbial functional genes related to oxidative phosphorylation compared with healthy controls. Butyrate and valerate levels were lower in transient AD infants compared with healthy and persistent AD infants.
CONCLUSIONS
Compositions, functions and metabolites of the early gut microbiome are related to natural courses of AD in infants.
MeSH Terms
Figure
Reference
-
1. Trompette A, Gollwitzer ES, Yadava K, Sichelstiel AK, Sprenger N, Ngom-Bru C, et al. Gut microbiota metabolism of dietary fiber influences allergic airway disease and hematopoiesis. Nat Med. 2014; 20:159–166.
Article2. Palm NW, de Zoete MR, Flavell RA. Immune-microbiota interactions in health and disease. Clin Immunol. 2015; 159:122–127.
Article3. Strachan DP. Hay fever, hygiene, and household size. BMJ. 1989; 299:1259–1260.
Article4. Platts-Mills TA. The allergy epidemics: 1870–2010. J Allergy Clin Immunol. 2015; 136:3–13.
Article5. Laughter D, Istvan JA, Tofte SJ, Hanifin JM. The prevalence of atopic dermatitis in Oregon schoolchildren. J Am Acad Dermatol. 2000; 43:649–655.
Article6. Spergel JM. From atopic dermatitis to asthma: the atopic march. Ann Allergy Asthma Immunol. 2010; 105:99–106.
Article7. Carlsten C, Dimich-Ward H, Ferguson A, Watson W, Rousseau R, Dybuncio A, et al. Atopic dermatitis in a high-risk cohort: natural history, associated allergic outcomes, and risk factors. Ann Allergy Asthma Immunol. 2013; 110:24–28.
Article8. Tham EH, Leung DY. Mechanisms by which atopic dermatitis predisposes to food allergy and the atopic march. Allergy Asthma Immunol Res. 2019; 11:4–15.
Article9. Abrahamsson TR, Jakobsson HE, Andersson AF, Bjorksten B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012; 129:434–440. 440.e1–432.
Article10. Lee E, Lee SY, Kang MJ, Kim K, Won S, Kim BJ, et al. Clostridia in the gut and onset of atopic dermatitis via eosinophilic inflammation. Ann Allergy Asthma Immunol. 2016; 117:91–92.e1.11. Lee MJ, Kang MJ, Lee SY, Lee E, Kim K, Won S, et al. Perturbations of gut microbiome genes in infants with atopic dermatitis according to feeding type. J Allergy Clin Immunol. 2018; 141:1310–1319.
Article12. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013; 341:569–573.
Article13. Zeng MY, Inohara N, Nuñez G. Mechanisms of inflammation-driven bacterial dysbiosis in the gut. Mucosal Immunol. 2017; 10:18–26.
Article14. Kim HK, Rutten NB, Besseling-van der Vaart I, Niers LE, Choi YH, Rijkers GT, et al. Probiotic supplementation influences faecal short chain fatty acids in infants at high risk for eczema. Benef Microbes. 2015; 6:783–790.
Article15. Lee SY, Lee E, Park YM, Hong SJ. Microbiome in the gut-skin axis in atopic dermatitis. Allergy Asthma Immunol Res. 2018; 10:354–362.
Article16. Nylund L, Nermes M, Isolauri E, Salminen S, de Vos WM, Satokari R. Severity of atopic disease inversely correlates with intestinal microbiota diversity and butyrate-producing bacteria. Allergy. 2015; 70:241–244.
Article17. Yang HJ, Lee SY, Suh DI, Shin YH, Kim BJ, Seo JH, et al. The Cohort for Childhood Origin of Asthma and allergic diseases (COCOA) study: design, rationale and methods. BMC Pulm Med. 2014; 14:109.
Article18. Hanifin JM, Rajka G. Diagnostic features of atopic dermatitis. Acta Derm Venereol. 1980; 60:44–47.19. Benjamini YH, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995; 57:289–300.
Article20. Penders J, Stobberingh EE, Thijs C, Adams H, Vink C, van Ree R, et al. Molecular fingerprinting of the intestinal microbiota of infants in whom atopic eczema was or was not developing. Clin Exp Allergy. 2006; 36:1602–1608.
Article21. Fujimura KE, Sitarik AR, Havstad S, Lin DL, Levan S, Fadrosh D, et al. Neonatal gut microbiota associates with childhood multisensitized atopy and T cell differentiation. Nat Med. 2016; 22:1187–1191.
Article22. van Nimwegen FA, Penders J, Stobberingh EE, Postma DS, Koppelman GH, Kerkhof M, et al. Mode and place of delivery, gastrointestinal microbiota, and their influence on asthma and atopy. J Allergy Clin Immunol. 2011; 128:948–955.e1-3.
Article23. Stiemsma LT, Arrieta MC, Dimitriu PA, Cheng J, Thorson L, Lefebvre DL, et al. Shifts in Lachnospira and Clostridium sp. in the 3-month stool microbiome are associated with preschool age asthma. Clin Sci (Lond). 2016; 130:2199–2207.24. Sjögren YM, Jenmalm MC, Böttcher MF, Björkstén B, Sverremark-Ekström E. Altered early infant gut microbiota in children developing allergy up to 5 years of age. Clin Exp Allergy. 2009; 39:518–526.
Article25. Sutherland DB, Fagarasan S. Gut reactions: eosinophils add another string to their bow. Immunity. 2014; 40:455–457.
Article26. Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011; 331:337–341.27. Reis RS, Horn F. Enteropathogenic Escherichia coli, Samonella, Shigella and Yersinia: cellular aspects of host-bacteria interactions in enteric diseases. Gut Pathog. 2010; 2:8.28. Wen H, Yin X, Yuan Z, Wang X, Su S. Comparative analysis of gut microbial communities in children under 5 years old with diarrhea. J Microbiol Biotechnol. 2018; 28:652–662.
Article29. Koropatkin NM, Cameron EA, Martens EC. How glycan metabolism shapes the human gut microbiota. Nat Rev Microbiol. 2012; 10:323–335.
Article30. Shin NR, Lee JC, Lee HY, Kim MS, Whon TW, Lee MS, et al. An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut. 2014; 63:727–735.31. Fieten KB, Totté JE, Levin E, Reyman M, Meijer Y, Knulst A, et al. Fecal microbiome and food allergy in pediatric atopic dermatitis: a cross-sectional pilot study. Int Arch Allergy Immunol. 2018; 175:77–84.
Article32. Roshanravan N, Mahdavi R, Alizadeh E, Ghavami A, Rahbar Saadat Y, Mesri Alamdari N, et al. The effects of sodium butyrate and inulin supplementation on angiotensin signaling pathway via promotion of Akkermansia muciniphila abundance in type 2 diabetes; a randomized, double-blind, placebo-controlled trial. J Cardiovasc Thorac Res. 2017; 9:183–190.33. Greer RL, Dong X, Moraes AC, Zielke RA, Fernandes GR, Peremyslova E, et al. Akkermansia muciniphila mediates negative effects of IFNγ on glucose metabolism. Nat Commun. 2016; 7:13329.
Article34. Ottman N, Reunanen J, Meijerink M, Pietilä TE, Kainulainen V, Klievink J, et al. Pili-like proteins of Akkermansia muciniphila modulate host immune responses and gut barrier function. PLoS One. 2017; 12:e0173004.35. Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2016; 137:852–860.36. Angelin A, Gil-de-Gómez L, Dahiya S, Jiao J, Guo L, Levine MH, et al. Foxp3 reprograms T cell metabolism to function in low-glucose, high-lactate environments. Cell Metab. 2017; 25:1282–1293.e7.
Article37. Hall A, Versalovic J. Microbial metabolism in the mammalian gut: molecular mechanisms and clinical implications. J Pediatr Gastroenterol Nutr. 2018; 66:Suppl 3. S72–S79.
Article38. De Vadder F, Kovatcheva-Datchary P, Goncalves D, Vinera J, Zitoun C, Duchampt A, et al. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell. 2014; 156:84–96.
Article39. Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, Yu D, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009; 461:1282–1286.
Article40. Kim M, Qie Y, Park J, Kim CH. Gut microbial metabolites fuel host antibody responses. Cell Host Microbe. 2016; 20:202–214.
Article41. Hong J, Jia Y, Pan S, Jia L, Li H, Han Z, et al. Butyrate alleviates high fat diet-induced obesity through activation of adiponectin-mediated pathway and stimulation of mitochondrial function in the skeletal muscle of mice. Oncotarget. 2016; 7:56071–56082.
Article42. Thorburn AN, McKenzie CI, Shen S, Stanley D, Macia L, Mason LJ, et al. Evidence that asthma is a developmental origin disease influenced by maternal diet and bacterial metabolites. Nat Commun. 2015; 6:7320.
Article43. Samuel BS, Shaito A, Motoike T, Rey FE, Backhed F, Manchester JK, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc Natl Acad Sci U S A. 2008; 105:16767–16772.
Article44. Payahoo L, Khajebishak Y, Alivand MR, Soleimanzade H, Alipour S, Barzegari A, et al. Investigation the effect of oleoylethanolamide supplementation on the abundance of Akkermansia muciniphila bacterium and the dietary intakes in people with obesity: a randomized clinical trial. Appetite. 2019; 141:104301.45. Lukovac S, Belzer C, Pellis L, Keijser BJ, de Vos WM, Montijn RC, et al. Differential modulation by Akkermansia muciniphila and Faecalibacterium prausnitzii of host peripheral lipid metabolism and histone acetylation in mouse gut organoids. MBio. 2014; 5:e01438-14.
Article46. Ottman N, Geerlings SY, Aalvink S, de Vos WM, Belzer C. Action and function of Akkermansia muciniphila in microbiome ecology, health and disease. Best Pract Res Clin Gastroenterol. 2017; 31:637–642.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Measurement of Atopic Dermatitis Disability
- Serum IgE Level in Patients of Atopic Dermatitis and Atopic Dermatitis with Molluscum Contagiosum
- Natural History and Risk Factors of Atopic Dermatitis in Children
- A Study of Natural Killer Cell Activity and Antibody - Dependent Cellular Cytotoxicity in Atopic Dermatitis
- Nipple Involvement in Atopic Dermatitis: Report of 3 cases