J Breast Cancer.
2018 Sep;21(3):277-287. 10.4048/jbc.2018.21.e39.
Long Noncoding RNA Signature and Disease Outcome in Estrogen Receptor-Positive Breast Cancer Patients Treated with Tamoxifen
- Affiliations
-
- 1Comprehensive Breast Health Center, Ruijin Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China. kwshen@medmail.com.cn
- KMID: 2421368
- DOI: http://doi.org/10.4048/jbc.2018.21.e39
Abstract
- PURPOSE
Recent data have shown that the expression levels of long noncoding RNAs (lncRNAs) are associated with tamoxifen sensitivity in estrogen receptor (ER)-positive breast cancer. Herein, we constructed an lncRNA-based model to predict disease outcomes of ER-positive breast cancer patients treated with tamoxifen.
METHODS
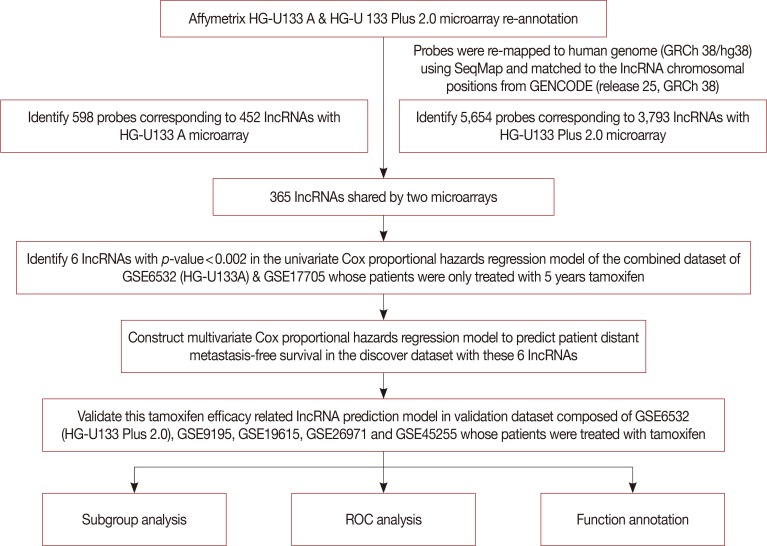
LncRNA expression information was acquired from Gene Expression Omnibus by re-mapping pre-existing microarrays of patients with ER-positive breast cancer treated with tamoxifen. The distant metastasis-free survival (DMFS) predictive signature was subsequently built based on a Cox proportional hazard regression model in discover cohort patients, which was further evaluated in another independent validation dataset.
RESULTS
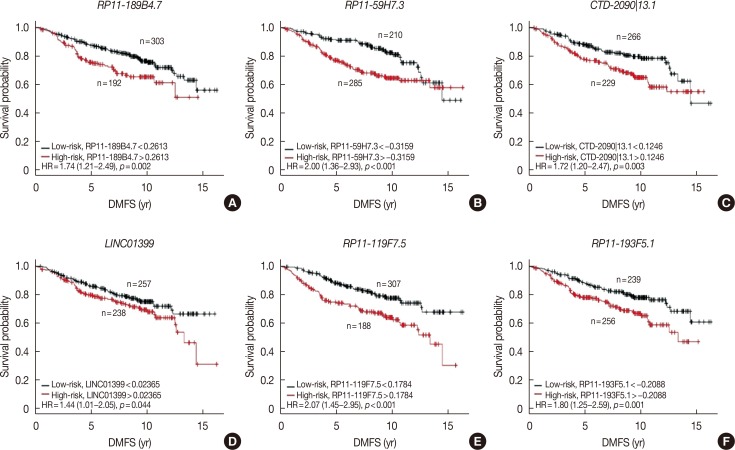
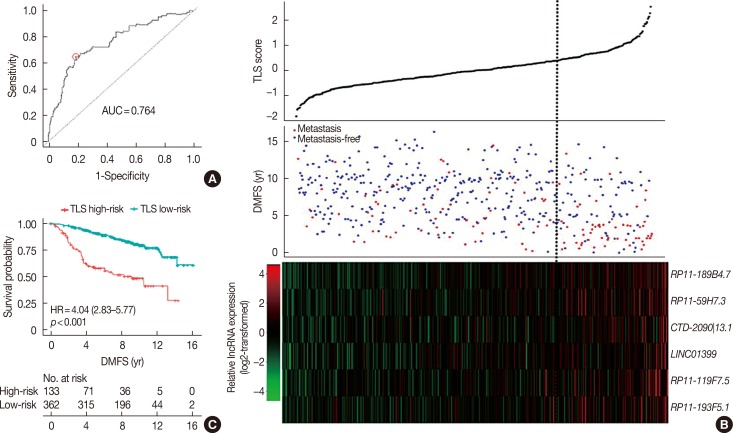
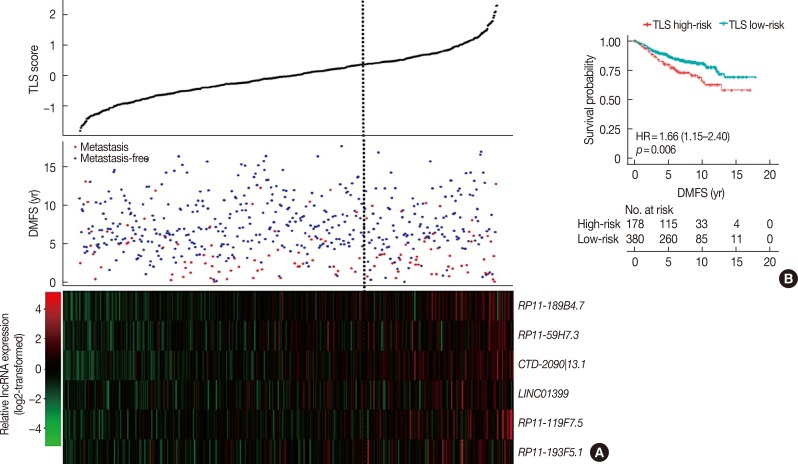
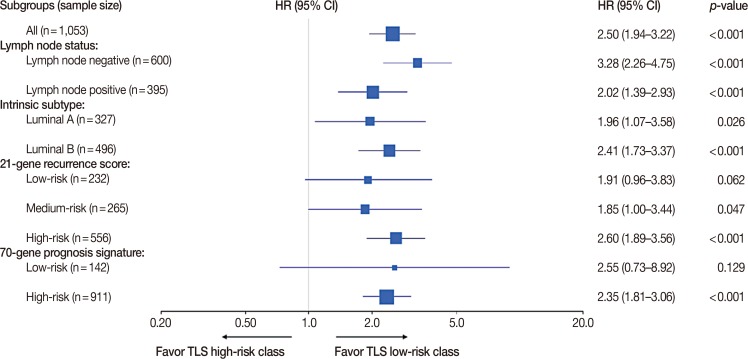
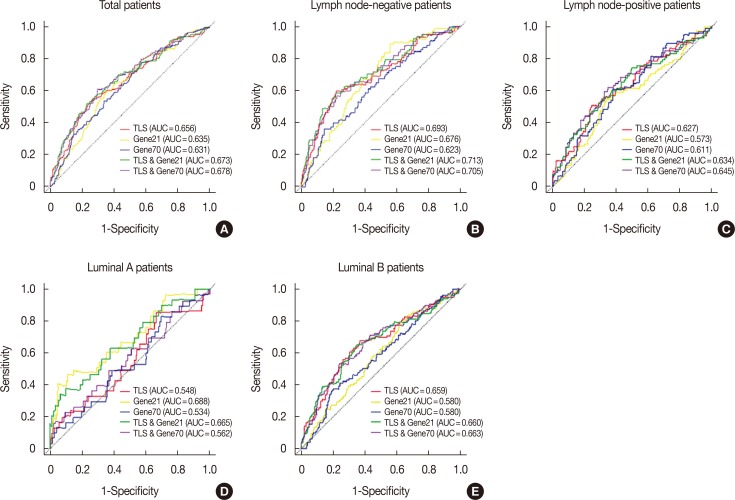
Six lncRNAs were found to be associated with DMFS in the discover cohort, which were used to construct a tamoxifen efficacy-related lncRNA signature (TLS). There were 133 and 362 patients with TLS high- and low-risk signatures in the discover cohort. Both univariate and multivariate analysis demonstrated that TLS was associated with DMFS. TLS high-risk patients had worse outcomes than low-risk patients, with a hazard ratio of 4.04 (95% confidence interval, 2.83-5.77; p < 0.001). Both subgroup analysis and receiver operating characteristic analysis indicated that TLS performed better in lymph node-negative, luminal B, 21-gene recurrence score high-risk, and 70-gene prognosis signature high-risk patients. Moreover, in a comparison of the 21-gene recurrence score and 70-gene prognosis signature, TLS showed a similar area under receiver operating characteristic curve in all patients. Gene Set Enrichment Analysis indicated that TLS high-risk patients showed different gene expression patterns related to the cell cycle and nucleotide metabolism from those of low-risk patients.
CONCLUSION
This six-lncRNA signature was associated with disease outcome in ER-positive breast cancer patients treated with tamoxifen, which is comparable to previous messenger RNA signatures and requires further clinical evaluation.
MeSH Terms
Figure
Reference
-
2. Montemurro F, Aglietta M. Hormone receptor-positive early breast cancer: controversies in the use of adjuvant chemotherapy. Endocr Relat Cancer. 2009; 16:1091–1102. PMID: 19726539.
Article3. Early Breast Cancer Trialists' Collaborative Group (EBCTCG). Effects of chemotherapy and hormonal therapy for early breast cancer on recurrence and 15-year survival: an overview of the randomised trials. Lancet. 2005; 365:1687–1717. PMID: 15894097.4. Vendrell JA, Ghayad S, Ben-Larbi S, Dumontet C, Mechti N, Cohen PA. A20/TNFAIP3, a new estrogen-regulated gene that confers tamoxifen resistance in breast cancer cells. Oncogene. 2007; 26:4656–4667. PMID: 17297453.
Article5. Jirström K, Rydén L, Anagnostaki L, Nordenskjöld B, Stål O, Thorstenson S, et al. Pathology parameters and adjuvant tamoxifen response in a randomised premenopausal breast cancer trial. J Clin Pathol. 2005; 58:1135–1142. PMID: 16254100.6. Kittaneh M, Montero AJ, Glück S. Molecular profiling for breast cancer: a comprehensive review. Biomark Cancer. 2013; 5:61–70. PMID: 24250234.
Article7. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006; 15 Spec No 1:R17–R29. PMID: 16651366.
Article8. Wang H, Guan Z, He K, Qian J, Cao J, Teng L. LncRNA UCA1 in anti-cancer drug resistance. Oncotarget. 2017; 8:64638–64650. PMID: 28969100.
Article9. Niknafs YS, Han S, Ma T, Speers C, Zhang C, Wilder-Romans K, et al. The lncRNA landscape of breast cancer reveals a role for DSCAM-AS1 in breast cancer progression. Nat Commun. 2016; 7:12791. PMID: 27666543.
Article10. Xue X, Yang YA, Zhang A, Fong KW, Kim J, Song B, et al. LncRNA HOTAIR enhances ER signaling and confers tamoxifen resistance in breast cancer. Oncogene. 2016; 35:2746–2755. PMID: 26364613.
Article11. Wang G, Chen X, Liang Y, Wang W, Shen K. A long noncoding RNA signature that predicts pathological complete remission rate sensitively in neoadjuvant treatment of breast cancer. Transl Oncol. 2017; 10:988–997. PMID: 29096247.
Article12. Du Z, Fei T, Verhaak RG, Su Z, Zhang Y, Brown M, et al. Integrative genomic analyses reveal clinically relevant long noncoding RNAs in human cancer. Nat Struct Mol Biol. 2013; 20:908–913. PMID: 23728290.
Article13. Zhou M, Zhong L, Xu W, Sun Y, Zhang Z, Zhao H, et al. Discovery of potential prognostic long non-coding RNA biomarkers for predicting the risk of tumor recurrence of breast cancer patients. Sci Rep. 2016; 6:31038. PMID: 27503456.
Article14. Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy: analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004; 20:307–315. PMID: 14960456.15. Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007; 8:118–127. PMID: 16632515.
Article16. Reese SE, Archer KJ, Therneau TM, Atkinson EJ, Vachon CM, de Andrade M, et al. A new statistic for identifying batch effects in highthroughput genomic data that uses guided principal component analysis. Bioinformatics. 2013; 29:2877–2883. PMID: 23958724.
Article17. Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008; 24:2395–2396. PMID: 18697769.
Article18. Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012; 22:1760–1774. PMID: 22955987.
Article19. Budczies J, Klauschen F, Sinn BV, Győrffy B, Schmitt WD, Darb-Esfahani S, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012; 7:e51862. PMID: 23251644.
Article20. Heagerty PJ, Lumley T, Pepe MS. Time-dependent ROC curves for censored survival data and a diagnostic marker. Biometrics. 2000; 56:337–344. PMID: 10877287.
Article21. Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005; 102:15545–15550. PMID: 16199517.
Article22. Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003; 34:267–273. PMID: 12808457.23. Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011; 39:3864–3878. PMID: 21247874.
Article24. Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009; 4:44–57. PMID: 19131956.
Article25. Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009; 37:1–13. PMID: 19033363.26. Merico D, Isserlin R, Stueker O, Emili A, Bader GD. Enrichment map: a network-based method for gene-set enrichment visualization and interpretation. PLoS One. 2010; 5:e13984. PMID: 21085593.
Article27. Gendoo DM, Ratanasirigulchai N, Schröder MS, Paré L, Parker JS, Prat A, et al. Genefu: an R/Bioconductor package for computation of gene expression-based signatures in breast cancer. Bioinformatics. 2016; 32:1097–1099. PMID: 26607490.28. Hayes EL, Lewis-Wambi JS. Mechanisms of endocrine resistance in breast cancer: an overview of the proposed roles of noncoding RNA. Breast Cancer Res. 2015; 17:40. PMID: 25849966.
Article29. Osborne CK, Boldt DH, Clark GM, Trent JM. Effects of tamoxifen on human breast cancer cell cycle kinetics: accumulation of cells in early G1 phase. Cancer Res. 1983; 43:3583–3585. PMID: 6861130.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Role of Long Noncoding RNAs in Antiestrogen Resistance in Breast Cancer: An Overview and Update
- Tamoxifen Resistance and Crosstalk of Signal Transduction in Breast Cancer
- The Effect of Tamoxifen of the Estrogen Receptor cDNA-Iipofected MDA-MB-231 Human Breast Cancer Cells
- Epigenetic Silencing of MORT Is an Early Event in Cancer and Is Associated with Luminal, Receptor Positive Breast Tumor Subtypes
- Restoration of Hormone Dependency in Estrogen Receptor - Lipofected MDA-MB-231 Human Breast Cancer Cells