J Korean Diabetes Assoc.
2007 Jul;31(4):326-335. 10.4093/jkda.2007.31.4.326.
Transcriptional Regulation of Insulin and CXCL10 Gene by Peroxisome Proliferator Activated Receptor gamma Coactivator-1alpha
- Affiliations
-
- 1Department of Internal Medicine, Kyungpook National University School of Medicine.
- 2Department of Genetic Engineering, Kyungpook National University.
- 3Department of Infectious Diseases, Keimyung University School of Medicine.
- KMID: 2222520
- DOI: http://doi.org/10.4093/jkda.2007.31.4.326
Abstract
-
BACKGROUND: Peroxisome proliferator-activated receptor-gamma coactivator 1alpha (PGC-1alpha), which act as a coactivator of nuclear receptors and several other transcription factors. This study was performed to evaluate the expressional regulation of insulin and inflammatory response genes by PGC-1alpha.
METHODS
Transient transfection assays were performed to measure the promoter activity of the insulin and CXCL10 gene. The insulin gene expression levels in INS-1 cells were determined by Northern blot analysis. Differentially expressed genes by PGC-1alpha overexpression in HASMCs were confirmed using DNA microarray, real-time PCR and Northen blot analysis.
RESULTS
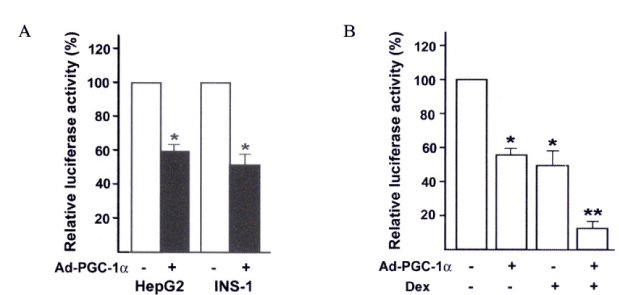
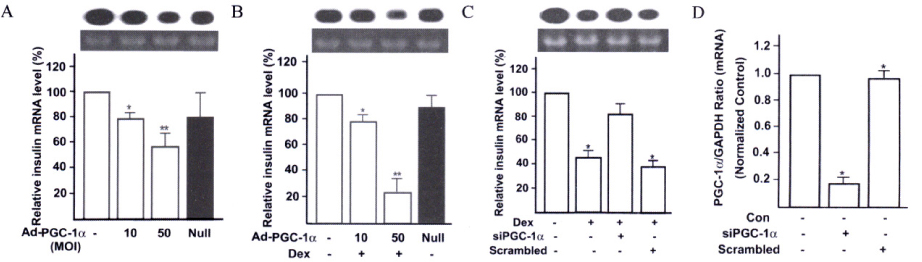
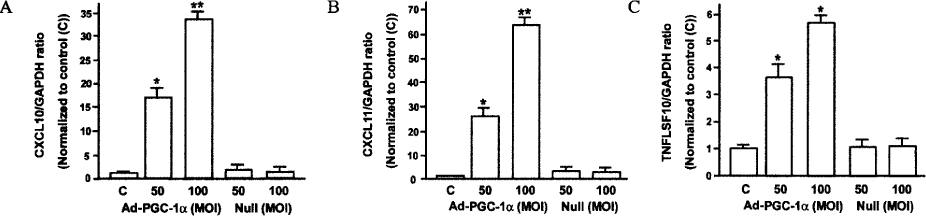
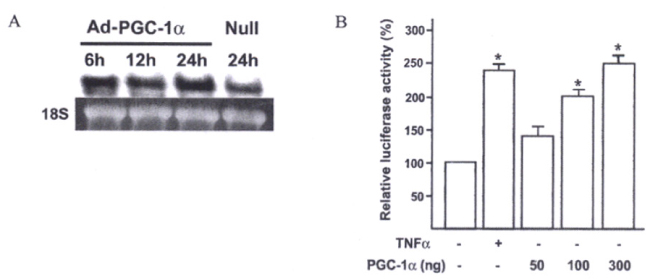
Insulin promoter activity and mRNA levels were suppressed by GR and Ad-PGC-1alpha. Northern blot analysis of the INS-1 cells revealed that infection with Ad-PGC-1alpha markedly reduced the amount of insulin mRNA and treatment of Dex enhanced this effect in an additive manner. The PGC-1alpha-specific siRNA decreased insulin expression that was induced by Dex in the GR-expressing INS-1 cells was nearly restored by this siRNA treatment. We found that when vascular smooth muscle cells (VSMCs) overexpressed PGC-1alpha, immune or inflammatory response genes were highly expressed. For example, promoter activity and mRNA level of CXCL10 gene were increased by PGC-1alpha.
CONCLUSION
PGC-1alpha overexpression inhibited insulin promoter activity in INS-1 cells and enhanced expressions of inflammatory response genes (CXCL10, CXCL11, TNFLSF10) in VSMCs.
Keyword
MeSH Terms
-
Blotting, Northern
Gene Expression
Insulin*
Muscle, Smooth, Vascular
Oligonucleotide Array Sequence Analysis
Peroxisomes*
PPAR gamma*
Real-Time Polymerase Chain Reaction
Receptors, Cytoplasmic and Nuclear
RNA, Messenger
RNA, Small Interfering
Transcription Factors
Transfection
Insulin
PPAR gamma
RNA, Messenger
RNA, Small Interfering
Receptors, Cytoplasmic and Nuclear
Transcription Factors
Figure
Reference
-
1. Melloul D, Marshak S, Cerasi E. Regulation of insulin gene transcription. Diabetologia. 2002. 45:309–326.2. Whelan J, Cordle SR, Henderson E, Weil PA, Stein R. Identification of a pancreatic beta-cell insulin gene transcription factor that binds to and appears to activate cell-type-specific expression: its possible relationship to other cellular factors that bind to a common insulin gene sequence. Mol Cell Biol. 1990. 10:1564–1572.3. Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993. 12:4251–4259.4. Peshavaria M, Gamer L, Henderson E, Teitelman G, Wright CV, Stein R. XIHbox 8, an endoderm-specific Xenopus homeodomain protein, is closely related to a mammalian insulin gene transcription factor. Mol Endocrinol. 1994. 8:806–816.5. Petersen HV, Serup P, Leonard J, Michelsen BK, Madesen OD. Transcriptional regulation of the human insulin gene is dependent on the homeodomain protein STF1/IPF1 acting through the CT boxes. Proc Natl Acad Sci USA. 1994. 91:10465–10469.6. Naya FJ, Stellrecht CM, Tsai MJ. Tissue-specific regulation of the insulin gene by a novel basic helix-loop-helix transcription factor. Genes Dev. 1995. 9:1009–1019.7. Olbrot M, Rud J, Moss LG, Sharma A. Identification of beta-cell-specific insulin gene transcription factor RIPE3b1 as mammalian MafA. Proc Nat Acad Sci USA. 2002. 99:6737–6742.8. Kataoka K, Han SI, Shioda S, Hirai M, Nishizawa M, Madsen OD. MafA is a glucose-regulated and pancreatic beta-cell-specific transcriptional activator for the insulin gene. J Biol Chem. 2002. 277:49903–49910.9. Matsuoka TA, Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Members of the large Maf transcription family regulate insulin gene transcription in islet beta cells. Mol Cell Biol. 2003. 23:6049–6062.10. Boam DS, Clark AR, Docherty K. Positive and negative regulation of the human insulin gene by multiple trans-acting factors. J Biol Chem. 1990. 265:8285–8289.11. Holme I, Solberg LA, Weissfeld L, Helgeland A, Hjermann I, Leren P, Strong JP, Williams OD. Coronary risk factors and their pathway of action through coronary raised lesions, coronary stenoses and coronary death. Multivariate statistical analysis of an autopsy series: the Oslo Study. Am J Cardiol. 1985. 55:40–47.12. Martin MJ, Hulley SB, Browner WS, Kuller LH, Wentworth D. Serum cholesterol, blood pressure, and mortality: implications from a cohort of 361,662 men. Lancet. 1986. 2:933–936.13. Ross R. The pathogenesis of atherosclerosis: a perspective for the 1990s. Nature. 1993. 362:801–808.14. Coleman KR, Braden GA, Willingham MC, Sane DC. Vitaxin, a humanized monoclonal antibody to the vitronectin receptor (alphavbeta3), reduces neointimal hyperplasia and total vessel area after balloon injury in hypercholesterolemic rabbits. Circ Res. 1999. 84:1268–1276.15. Rollins BJ. Chemokines. Blood. 1997. 90:909–928.16. Umehara H, Bloom ET, Okazaki T, Nagano Y, Yoshie O, Imai T. Fractalkine in vascular biology: from basic research to clinical disease. Arterioscler Thromb Vasc Biol. 2004. 24:34–40.17. Schecter AD, Berman AB, Taubman MB. Chemokine receptors in vascular smooth muscle. Microcirculation. 2003. 10:265–272.18. Wang X, Yue TL, Ohlstein EH, Sung CP, Feuerstein GZ. Interferon-inducible protein-10 involves vascular smooth muscle cell migration, proliferation, and infla-mmatory response. J Biol Chem. 1996. 271:24286–24293.19. Brightling CE, Ammit AJ, Kaur D, Black JL, Wardlaw AJ, Hughes JM, Bradding P. The CXCL10/CXCR3 axis mediates human lung mast cell migration to asthmatic airway smooth muscle. Am J Respir Crit Care Med. 2005. 171:1103–1108.20. Puigserver P, Wu Z, Park CW, Graves R, Wright M, spiegelman BM. A cold-inducible coactivator of nuclear receptors linked to adaptive thermogenesis. Cell. 1998. 92:829–839.21. Vega RB, huss JM, Kelly DP. The coactivator PGC-1 cooperates with Peroxisome proliferator-activated receptor alpha in transcriptional control of nuclear genes encoding mitochondrial fatty acid oxidation enzymes. Mol Cell Biol. 2000. 20:1868–1876.22. Knutti D, Kaul A, Kralli A. A tissue-specific coactivator of steroid receptors, identified in a functional genetic screen. Mol Cell Biol. 2000. 20:2411–2422.23. Delerive P, Wu Y, Burris TP, Chin WW, Suen CS. PGC-1 functionsas a transcriptional coactivator for the retinoid X receptors. J Biol Chem. 2002. 277:3913–3917.24. Tcherepanova I, Puigserver P, Norris JD, Spiegelman BM, McDonnell DP. Modulation of estrogen receptor-alpha transcriptional activity by the coactivator PGC-1. J Biol Chem. 2000. 275:16302–16308.25. Oberkofler H, Schraml E, Krempler F, Patsch W. Potentiation of liver X receptpr transcriptional activity by peroxisome proliferators activated receptor g coactivator-1a. Biochem J. 2003. 371:89–96.26. Denis M, Gustafsson JA, Wikstrom AC. Interaction of the Mr=90,000 heat shock protein with the steroid-binding domain of the glucocorticoid receptor. J Biol Chem. 1988. 263:18520–18523.27. Wrange O, Okret S, Radojcic M, Carlstedt-Duke J, Gustafsson JA. Characterization of the purified activated glucocorticoid receptor from rat liver cytosol. J Biol Chem. 1984. 259:4534–4541.28. Bresnick EH, Dalman FC, Pratt WB. Direct stoichiometric evidence that the untransformed Mr 300,000,9S, glucocorticoid receptor is a core unit derived from a larger heteromeric complex. Biochemistry. 1990. 29:520–527.29. Scheidereit C, Geisse S, Westphal HM, Beato M. The glucocorticoid receptor binds to defined nucleotide sequences near the promoter of mouse mammary tumour virus. Nature. 1983. 304:749–752.30. Beato M. Gene regulation by steroid hormones. Cell. 1989. 56:335–343.31. Funder JW. Mineralocorticoids, glucocorticoids, receptors and response elements. Science. 1993. 259:1132–1133.32. Scott DK, Stromstedt PE, Wang JC, Granner DK. Further characterization of the glucocorticoid response unit in the phosphoenolpyruvate carboxykinase gene. The role of the glucocorticoid receptor-binding sites. Mol Endocrinol. 1998. 12:482–491.33. Jonat C, Rahmsdorf HJ, Park KK, Cato AC, Gebel S, Ponta H, Herrlich P. Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990. 62:1189–1204.34. Majumder S, Zhou LZ, Chaturvedi P, Babcock G, Aras S, Ransohoff RM. p48/STAT-1alpha-containing complexes play a predominant role in induction of IFN-gamma-inducible protein, 10 kDa (IP-10) by IFN-gamma alone or in synergy with TNF-alpha. J Immunol. 1998. 161:4736–4744.35. Okita K, Yang Q, Yamagata K, Hangenfeldt KA, Miyagawa J, Kajimoto Y, Nakajima H, Namba M, Wollheim CB, Hanafusa T, Matsuzawa Y. Human insulin gene is a target gene of hepatocyte nuclear factor-1alpha (HNF-1alpha) and HNF-1beta. Biochem Biophys Res Commun. 1999. 24:566–569.36. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)). Methods. 2001. 25:402–408.37. Ding WQ, Dong M, Ninova D, Holicky EL, Stegall MD, Miller LJ. Forskolin suppresses insulin gene transcription in islet beta-cells through a protein kinase A-independent pathway. Cell Signal. 2003. 15:27–33.38. Laubner K, Kieffer TJ, Lam NT, Niu X, Jakob F, Seufert J. Inhibition of preproinsulin gene expression by leptin induction of suppressor of cytokine signaling 3 in pancreatic beta-cells. Diabetes. 2005. 54:3410–3417.39. Olefsky JM, Johnson J, Liu F, Jen P, Reaven GM. The effects of acute and chronic dexamethasone administration on insulin binding to isolated rat hepatocytes and adipocytes. Metabolism. 1975. 24:517–527.40. Pierluissi J, Navas F, Ashcroft SJ. Effect of adrenal steroids on insulin release from cultured rat islets of Langerhans. Diabetologia. 1986. 29:119–121.41. Olefsky JM, Kimmerling G. Effects of glucocorticoids on carbohydrate metabolism. Am J Med Sci. 1976. 271:20–21.42. Schacke H, Docke WD, Asadullah K. Mechanisms involved in the side effects of glucocorticoids. Pharmacol Ther. 2002. 96:23–43.43. Fryer CJ, Nordeen SK, Archer TK. Antiprogestins mediate differential effects on glucocorticoid receptor remodeling of chromatin structure. J Biol Chem. 1998. 273:1175–1183.44. List HJ, Lozano C, Lu J, Danielsen M, Wellstein A, Riegel AT. Comparison of chromatin remodeling and transcriptional activation of the mouse mammary tumor virus promoter by the androgen and glucocorticoid receptor. Exp Cell Res. 1999. 250:414–422.45. Schoneveld OJ, Gaemers IC, Lamers WH. Mechanisms of glucocorticoid signaling. Biochim Biophys Acta. 2004. 1680:114–128.46. Kim HJ, Park KG, Yoo EK, Kim YH, Kim YN, Kim HS, Kim HT, Kim YH, Park JY, Lee KU, Jang WG, Kim JG, Kim BW, Lee IK. Effects of PGC-1α on TNF-α induced MCP-1 and VCAM-1 expression and NF-κB activation in human aortic smooth muscle and endothelial cells. Antioxidants & Redox Signaling. 2007. 9:301–307.47. St-Pierre J, Drori S, Uldry M, Silvaggi JM, Rhee J, Jager S, Handschin C, Zheng K, Lin J, Yang W, Simon DK, Bachoo R, Spiegelman BM. Suppression of reactive oxygen species and neurodegeneration by the PGC-1 transcriptional coactivators. Cell. 2006. 127:397–408.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Sterol-independent repression of low density lipoprotein receptor promoter by peroxisome proliferator activated receptor gamma coactivator-1alpha (PGC-1alpha)
- The Polymorphisms of PPAR-gamma Coactivator 1alpha Gly482Ser (PGC-1alpha Gly482Ser) are Associated with the Nephropathy of Korean Patients with Type 2 Diabetes Mellitus
- Effects of Sulfonylureas on Peroxisome Proliferator-Activated Receptor gamma Activity and on Glucose Uptake by Thiazolidinediones
- Peroxisome Proliferator Activated Receptor-delta (PPAR-delta)
- The association between PGC-1a and Alzheimer's disease