Intest Res.
2015 Jan;13(1):39-49. 10.5217/ir.2015.13.1.39.
Comparative Analysis of Gastrointestinal Microbiota Between Normal and Caudal-Related Homeobox 2 (Cdx2) Transgenic Mice
- Affiliations
-
- 1Division of Gastroenterology, Department of Medicine, Jichi Medical University, Shimotsuke, Japan. muto@jichi.ac.jp
- 2Yakult Honsha Co., Ltd., Yakult Central Institute, Kunitachi, Japan.
- 3Department of Microbiology, Kitasato University School of Medicine, Sagamihara, Japan.
- KMID: 1807375
- DOI: http://doi.org/10.5217/ir.2015.13.1.39
Abstract
- BACKGROUND/AIMS
Caudal-related homeobox 2 (Cdx2) is expressed in the human intestinal metaplastic mucosa and induces intestinal metaplastic mucosa in the Cdx2 transgenic mouse stomach. Atrophic gastritis and intestinal metaplasia commonly lead to gastric achlorhydria, which predisposes the stomach to bacterial overgrowth. In the present study, we determined the differences in gut microbiota between normal and Cdx2 transgenic mice, using quantitative reverse transcription-polymerase chain reaction (qRT-PCR).
METHODS
Twelve normal (control) and 12 Cdx2 transgenic mice were sacrificed, and the gastric, jejunal, ileac, cecal and colonic mucosa, and feces were collected. To quantitate bacterial microbiota, we used real-time qRTPCR with 16S rRNA gene-targeted, species-specific primers.
RESULTS
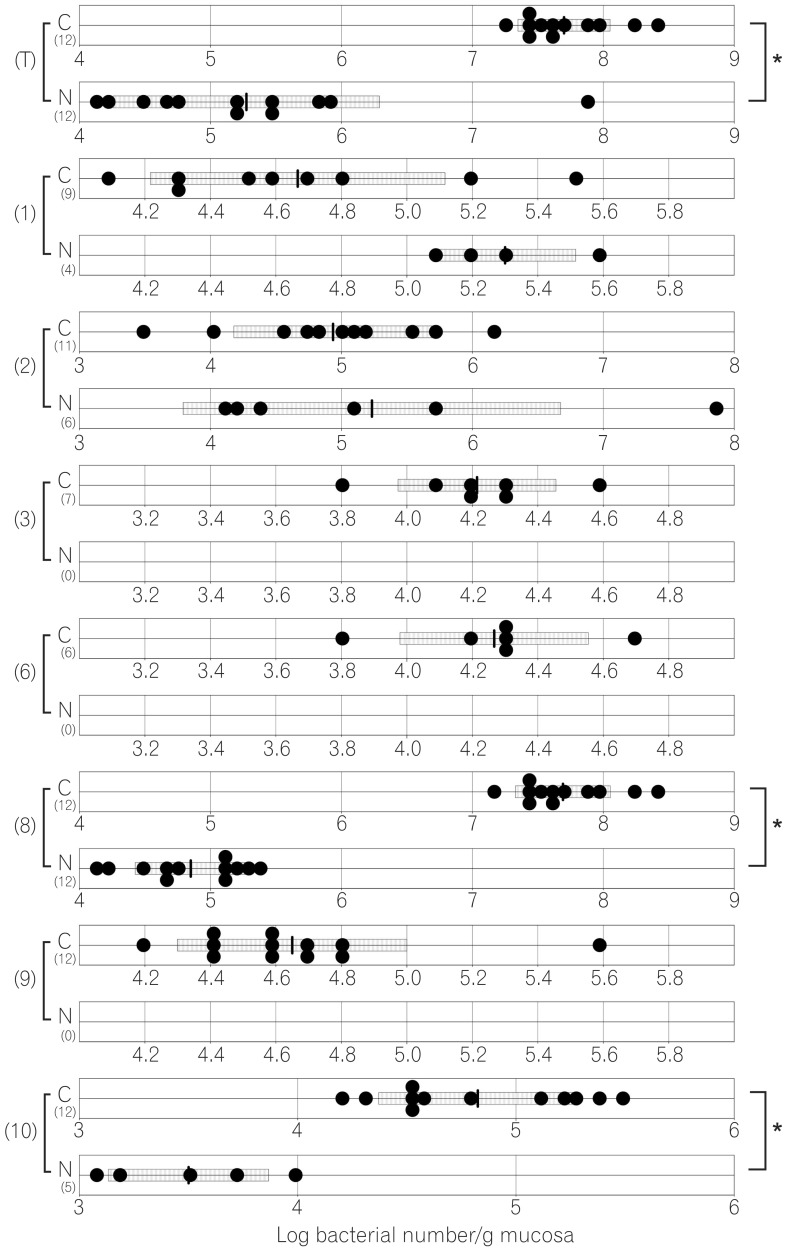
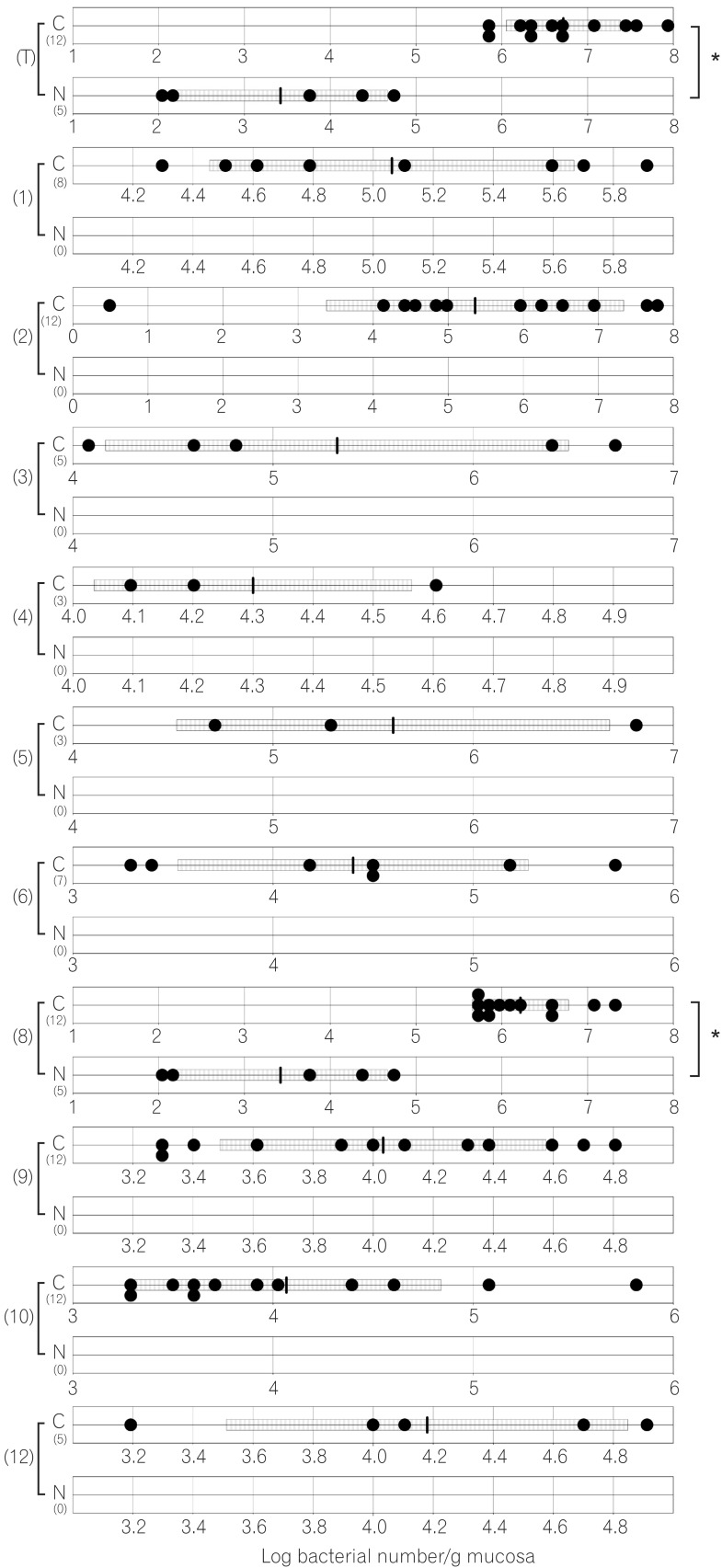
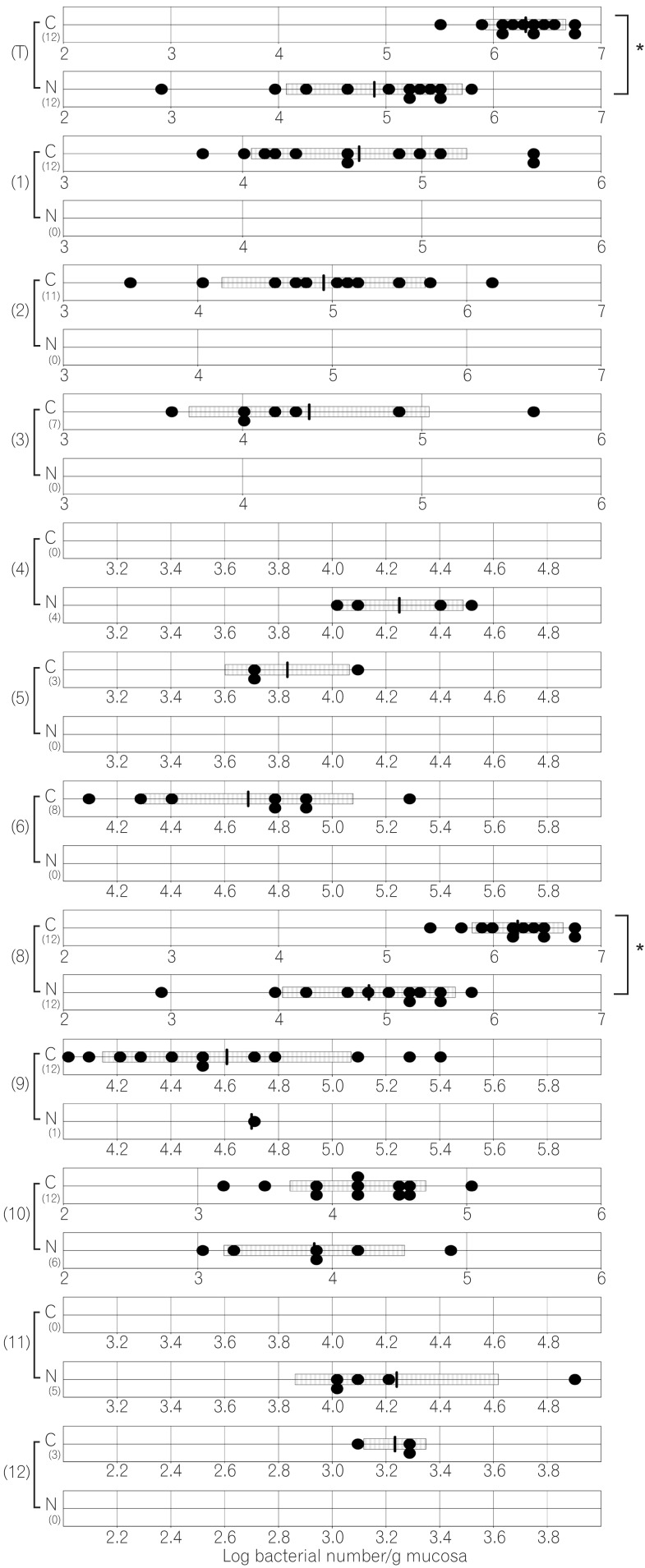
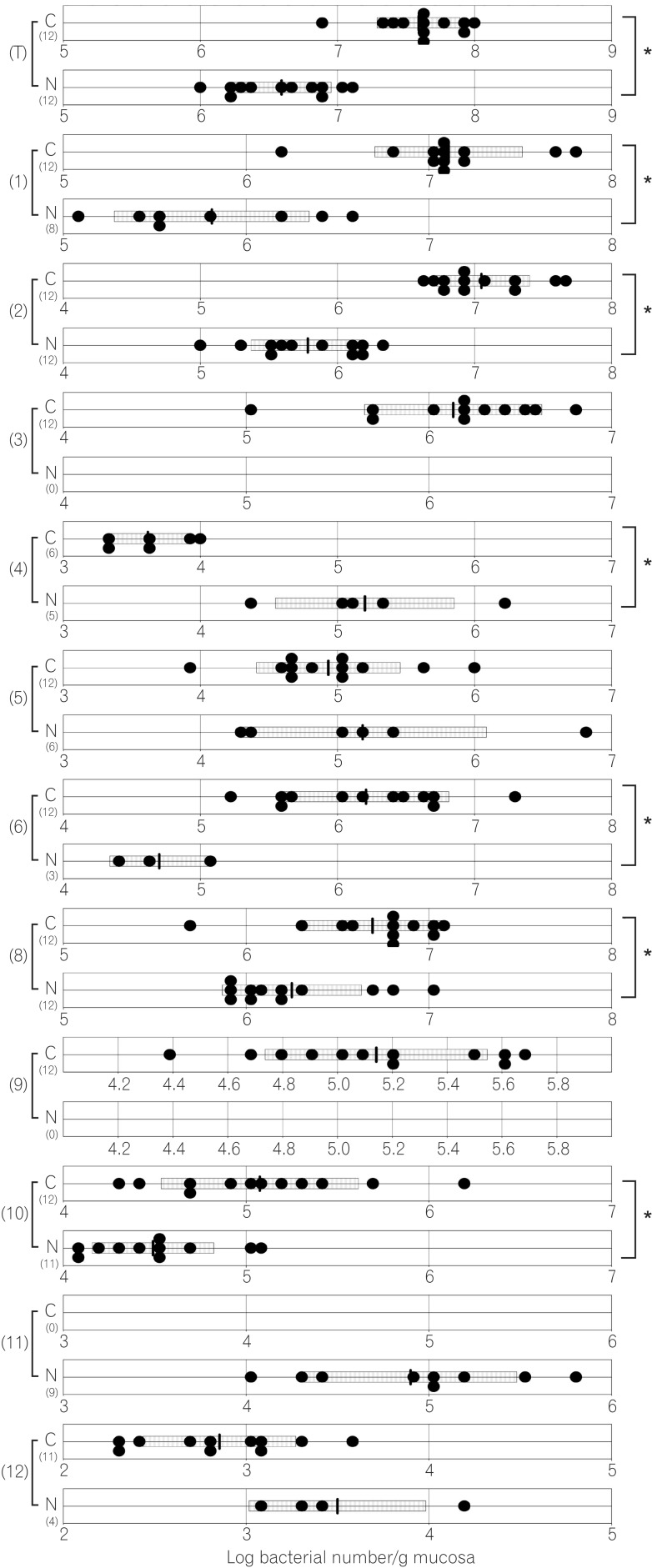
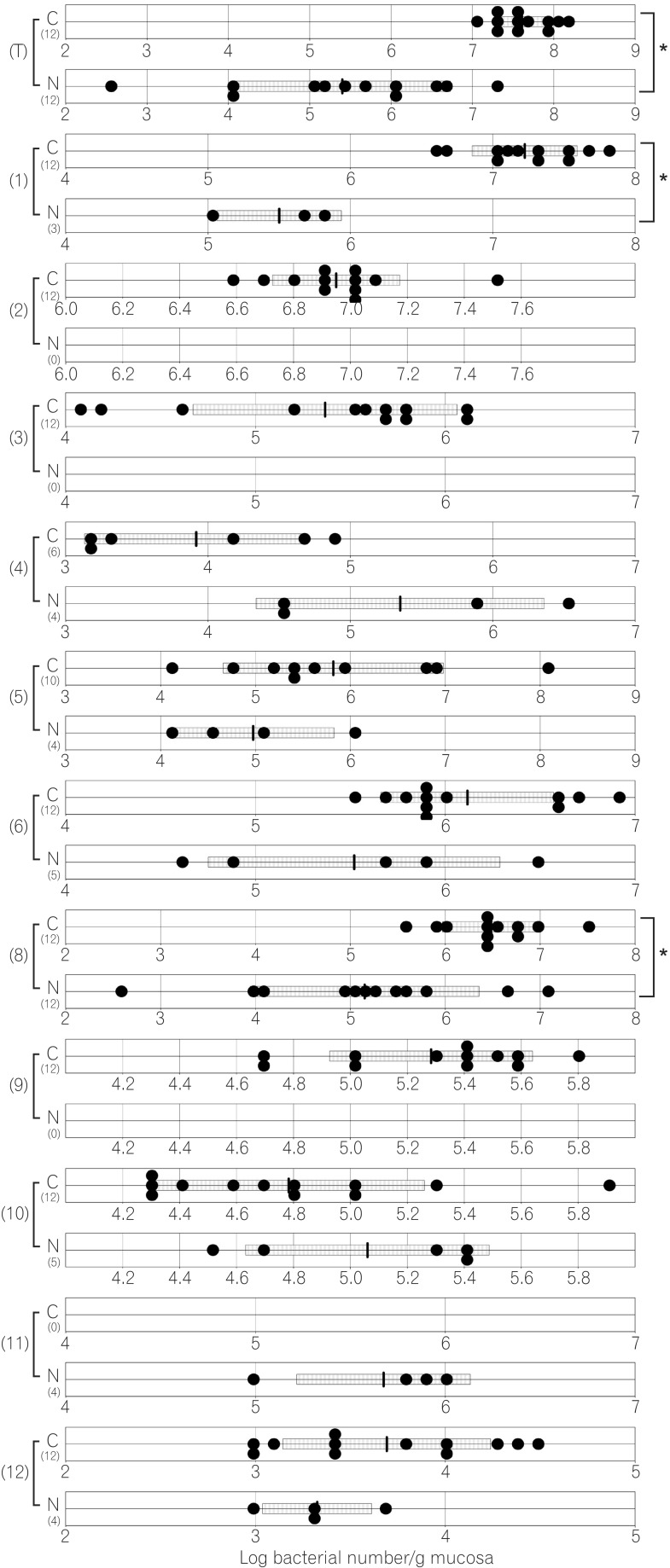
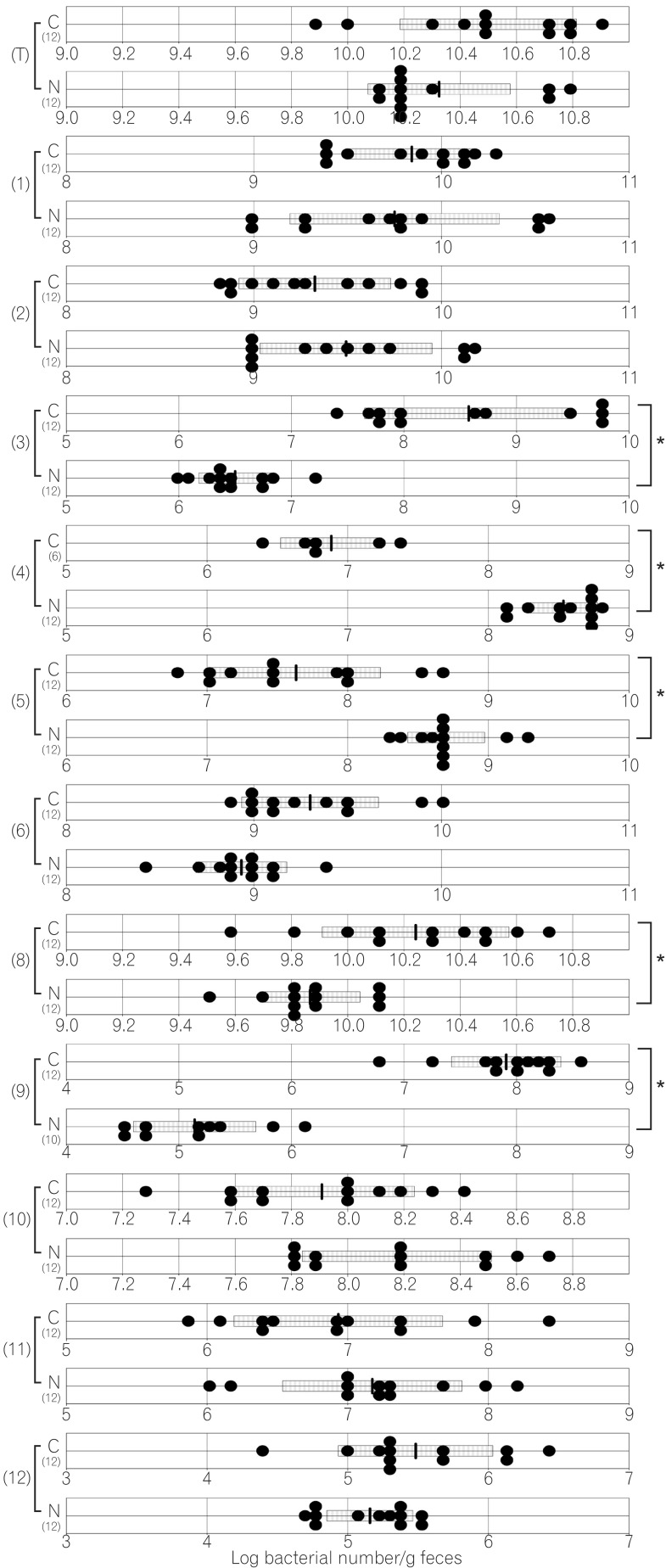
The total numbers of bacteria in the gastric, jejunal, ileac, cecal, and colonic mucosa of the Cdx2 transgenic mice were significantly higher than those of the normal mice. The Bacteroides fragilis group and also Prevotella were not detected in the stomach of the normal mice, although they were detected in the Cdx2 transgenic mice. Moreover, the Clostridium coccoides group, Clostridium leptum subgroup, Bacteroides fragilis group, and Prevotella were not detected in the jejunum or ileum of the normal mice, although they were detected in the Cdx2 transgenic mice. The fecal microbiota of the normal mice was similar to that of the Cdx2 transgenic mice.
CONCLUSIONS
Our results showed the differences in composition of gut microbiota between normal and Cdx2 transgenic mice, which may be caused by the development of gastric achlorhydria and intestinal metaplasia in Cdx2 transgenic mice.
Keyword
MeSH Terms
Figure
Reference
-
1. Meyer BI, Gruss P. Mouse Cdx-1 expression during gastrulation. Development. 1993; 117:191–203. PMID: 7900985.2. Suh E, Chen L, Taylor J, Traber PG. A homeodomain protein related to caudal regulates intestine-specific gene transcription. Mol Cell Biol. 1994; 14:7340–7351. PMID: 7935448.
Article3. Suh E, Traber PG. An intestine-specific homeobox gene regulates proliferation and differentiation. Mol Cell Biol. 1996; 16:619–625. PMID: 8552090.
Article4. Mizoshita T, Inada K, Tsukamoto T, et al. Expression of Cdx1 and Cdx2 mRNAs and relevance of this expression to differentiation in human gastrointestinal mucosa--with special emphasis on participation in intestinal metaplasia of the human stomach. Gastric Cancer. 2001; 4:185–191. PMID: 11846061.
Article5. Eda A, Osawa H, Yanaka I, et al. Expression of homeobox gene CDX2 precedes that of CDX1 during the progression of intestinal metaplasia. J Gastroenterol. 2002; 37:94–100. PMID: 11871772.
Article6. Satoh K, Mutoh H, Eda A, et al. Aberrant expression of CDX2 in the gastric mucosa with and without intestinal metaplasia: effect of eradication of Helicobacter pylori. Helicobacter. 2002; 7:192–198. PMID: 12047325.7. Bai YQ, Yamamoto H, Akiyama Y, et al. Ectopic expression of homeodomain protein CDX2 in intestinal metaplasia and carcinomas of the stomach. Cancer Lett. 2002; 176:47–55. PMID: 11790453.
Article8. Almeida R, Silva E, Santos-Silva F, et al. Expression of intestinespecific transcription factors, CDX1 and CDX2, in intestinal metaplasia and gastric carcinomas. J Pathol. 2003; 199:36–40. PMID: 12474224.
Article9. Mutoh H, Hakamata Y, Sato K, et al. Conversion of gastric mucosa to intestinal metaplasia in Cdx2-expressing transgenic mice. Biochem Biophys Res Commun. 2002; 294:470–479. PMID: 12051735.
Article10. Watanabe H. Intestinal metaplasia -the effect of Acid on the gastric mucosa and gastric carcinogenesis. J Toxicol Pathol. 2010; 23:115–123. PMID: 22272022.
Article11. Bik EM, Eckburg PB, Gill SR, et al. Molecular analysis of the bacterial microbiota in the human stomach. Proc Natl Acad Sci U S A. 2006; 103:732–737. PMID: 16407106.12. Maldonado-Contreras A, Goldfarb KC, Godoy-Vitorino F, et al. Structure of the human gastric bacterial community in relation to Helicobacter pylori status. ISME J. 2011; 5:574–579. PMID: 20927139.13. Osaki T, Matsuki T, Asahara T, et al. Comparative analysis of gastric bacterial microbiota in Mongolian gerbils after long-term infection with Helicobacter pylori. Microb Pathog. 2012; 53:12–18. PMID: 22783557.14. Suau A, Bonnet R, Sutren M, et al. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl Environ Microbiol. 1999; 65:4799–4807. PMID: 10543789.15. Langendijk PS, Schut F, Jansen GJ, et al. Quantitative fluorescence in situ hybridization of Bifidobacterium spp. with genusspecific 16S rRNA-targeted probes and its application in fecal samples. Appl Environ Microbiol. 1995; 61:3069–3075. PMID: 7487040.16. Satokari RM, Vaughan EE, Akkermans AD, Saarela M, De Vos WM. Polymerase chain reaction and denaturing gradient gel electrophoresis monitoring of fecal Bifidobacterium populations in a prebiotic and probiotic feeding trial. Syst Appl Microbiol. 2001; 24:227–231. PMID: 11518325.17. Matsuda K, Tsuji H, Asahara T, Matsumoto K, Takada T, Nomoto K. Establishment of an analytical system for the human fecal microbiota, based on reverse transcription-quantitative PCR targeting of multicopy rRNA molecules. Appl Environ Microbiol. 2009; 75:1961–1969. PMID: 19201979.18. Matsuda K, Tsuji H, Asahara T, Kado Y, Nomoto K. Sensitive quantitative detection of commensal bacteria by rRNA-targeted reverse transcription-PCR. Appl Environ Microbiol. 2007; 73:32–39. PMID: 17071791.19. Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol. 2004; 70:7220–7228. PMID: 15574920.20. Matsuki T. Development of quantitative PCR detection method with 16S rRNA gene-targeted genus- and species-specific primers for the analysis of human intestinal microflora and its application. Nihon Saikingaku Zasshi. 2007; 62:255–261. PMID: 17575792.21. Kikuchi E, Miyamoto Y, Narushima S, Itoh K. Design of speciesspecific primers to identify 13 species of Clostridium harbored in human intestinal tracts. Microbiol Immunol. 2002; 46:353–358. PMID: 12139395.22. Watanabe K. inventor. Yakult Honsha Co., Ltd., assignee. Primers for lactic acid bacterium. Japan patent 11-151097. 1999. 6. 08.23. Sakaguchi S, Saito M, Tsuji H, et al. Bacterial rRNA-targeted reverse transcription-PCR used to identify pathogens responsible for fever with neutropenia. J Clin Microbiol. 2010; 48:1624–1628. PMID: 20351213.24. Stearns JC, Lynch MD, Senadheera DB, et al. Bacterial biogeography of the human digestive tract. Sci Rep. 2011; 1:170. PMID: 22355685.25. Gronlund MM, Grzeskowiak L, Isolauri E, Salminen S. Influence of mother's intestinal microbiota on gut colonization in the infant. Gut Microbes. 2011; 2:227–233. PMID: 21983067.26. Gu S, Chen D, Zhang JN, et al. Bacterial community mapping of the mouse gastrointestinal tract. PLoS One. 2013; 8:e74957. PMID: 24116019.27. Mutoh H, Sakurai S, Satoh K, et al. Development of gastric carcinoma from intestinal metaplasia in Cdx2-transgenic mice. Cancer Res. 2004; 64:7740–7747. PMID: 15520178.28. Lofgren JL, Whary MT, Ge Z, et al. Lack of commensal flora in Helicobacter pylori-infected INS-GAS mice reduces gastritis and delays intraepithelial neoplasia. Gastroenterology. 2011; 140:210–220. PMID: 20950613.29. Dicksved J, Lindberg M, Rosenquist M, Enroth H, Jansson JK, Engstrand L. Molecular characterization of the stomach microbiota in patients with gastric cancer and in controls. J Med Microbiol. 2009; 58:509–516. PMID: 19273648.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Relationship of CDX1 mRNA and CDX2 mRNA expression and clinicopathologic features in colorectal cancers
- Cyclooxygenase 2 in Gastric Carcinoma Is Expressed in Doublecortin- and CaM Kinase-Like-1-Positive Tuft Cells
- Relationships Between the Expressions of CDX1 and CDX2 mRNA and Clinicopathologic Features in Colorectal Cancers
- Expression of Cdx2 Protein in Colorectal Cancer
- Positive Selection of Transgenic CD4 T Cells in the Restricted Peptide Environment