Yonsei Med J.
2013 May;54(3):772-777. 10.3349/ymj.2013.54.3.772.
Human SNF2L Gene Is Regulated Constitutively and Inducibly in Neural Cells via a cAMP-Response Element
- Affiliations
-
- 1Department of Center Laboratory, Provincial Hospital Affiliated to Shandong University, Jinan, China. yrzhao@sdu.edu.cn
- 2Key Laboratory for Experimental Teratology of the Ministry of Education and Institute of Medical Genetics, Shandong University School of Medicine, Jinan, China. xiaxixi716@yahoo.com.cn
- KMID: 1727896
- DOI: http://doi.org/10.3349/ymj.2013.54.3.772
Abstract
- PURPOSE
SNF2L belongs to Imitation Switch family and plays an essential role in neural tissues and gonads. In our previous studies, we have demonstrated that the basal transcription of human SNF2L gene is regulated by two cis-elements, cAMP response element (CRE)- and Sp1-binding sites. Recent studies suggested that cyclic adenosine monophosphate (cAMP) stimulation significantly up-regulated SNF2L expression in ovarian granulose cells. These data suggested that protein kinase-mediated signal pathways might also regulate SNF2L expression in neural cells. We therefore investigated the effects of agents that activate protein kinases A on SNF2L gene expression in neural cells.
MATERIALS AND METHODS
To increase intracellular cAMP levels, all neural cells were treated with forskolin and dbcAMP, two cAMP response activators. We exmined the effects of cAMP on the promoter activity of human SNF2L gene by luciferase reporter gene assays, and further examined the effects of cAMP on endogenous SNF2L mRNA levels by qPCR.
RESULTS
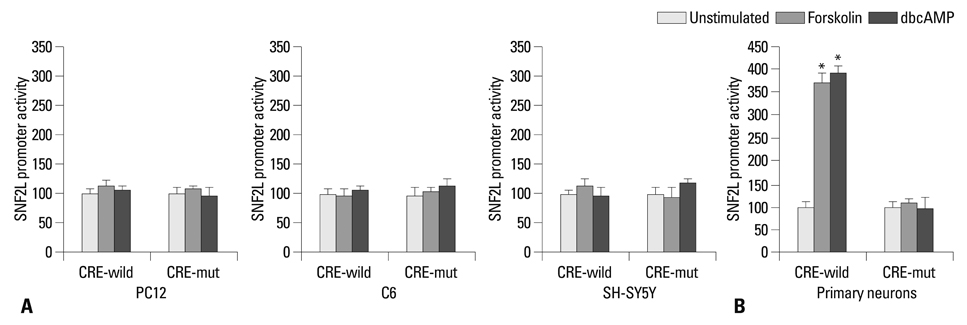
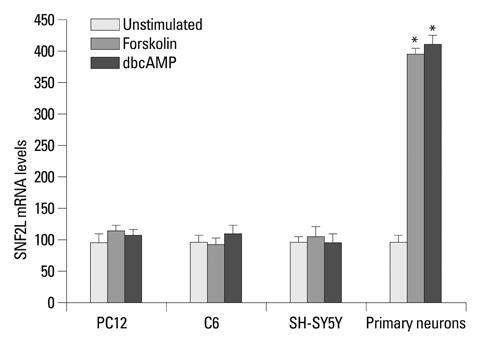
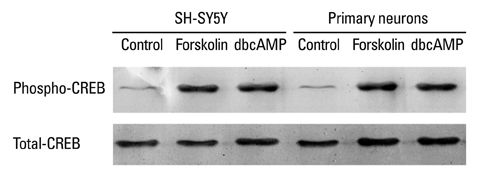
Transient expression of a luciferase fusion gene under the control of the SNF2L promoter was significantly increased by treatment of rat primary neurons with forskolin or dbcAMP, but not PC12, C6 and SH-SY5Y cells. Consistently, treatment with forskolin or dbcAMP could enhance endogenous SNF2L mRNA levels also only in rat primary neurons.
CONCLUSION
These results suggest that the CRE consensus sequence in the SNF2L proximal promoter most likely confers constitutive activation and regulation by cAMP in neural cells.
Keyword
MeSH Terms
-
Animals
Bucladesine/pharmacology
Cell Line
Colforsin/pharmacology
Cyclic AMP/*metabolism
DNA-Binding Proteins/chemistry/*genetics/metabolism
*Gene Expression Regulation
Humans
Luciferases/analysis
Neurons/*metabolism
PC12 Cells
Promoter Regions, Genetic
RNA, Messenger/metabolism
Rats
Rats, Wistar
Recombinant Fusion Proteins/analysis
*Response Elements
Transcription Factors/chemistry/*genetics/metabolism
DNA-Binding Proteins
RNA, Messenger
Recombinant Fusion Proteins
Transcription Factors
Colforsin
Bucladesine
Cyclic AMP
Luciferases
Figure
Reference
-
1. Hogan C, Varga-Weisz P. The regulation of ATP-dependent nucleosome remodelling factors. Mutat Res. 2007. 618:41–51.
Article2. Gangaraju VK, Bartholomew B. Mechanisms of ATP dependent chromatin remodeling. Mutat Res. 2007. 618:3–17.
Article3. Brown E, Malakar S, Krebs JE. How many remodelers does it take to make a brain? Diverse and cooperative roles of ATP-dependent chromatin-remodeling complexes in development. Biochem Cell Biol. 2007. 85:444–462.
Article4. Johnson CN, Adkins NL, Georgel P. Chromatin remodeling complexes: ATP-dependent machines in action. Biochem Cell Biol. 2005. 83:405–417.
Article5. Corona DF, Tamkun JW. Multiple roles for ISWI in transcription, chromosome organization and DNA replication. Biochim Biophys Acta. 2004. 1677:113–119.6. Lazzaro MA, Picketts DJ. Cloning and characterization of the murine Imitation Switch (ISWI) genes: differential expression patterns suggest distinct developmental roles for Snf2h and Snf2l. J Neurochem. 2001. 77:1145–1156.
Article7. Barak O, Lazzaro MA, Lane WS, Speicher DW, Picketts DJ, Shiekhattar R. Isolation of human NURF: a regulator of Engrailed gene expression. EMBO J. 2003. 22:6089–6100.
Article8. Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, et al. The Drosophila nucleosome remodeling factor NURF is required for Ecdysteroid signaling and metamorphosis. Genes Dev. 2005. 19:2540–2545.
Article9. Pépin D, Vanderhyden BC, Picketts DJ, Murphy BD. ISWI chromatin remodeling in ovarian somatic and germ cells: revenge of the NURFs. Trends Endocrinol Metab. 2007. 18:215–224.
Article10. Ye Y, Xiao Y, Wang W, Wang Q, Yearsley K, Wani AA, et al. Inhibition of expression of the chromatin remodeling gene, SNF2L, selectively leads to DNA damage, growth inhibition, and cancer cell death. Mol Cancer Res. 2009. 7:1984–1999.
Article11. Yip DJ, Corcoran CP, Alvarez-Saavedra M, DeMaria A, Rennick S, Mears AJ, et al. Snf2l regulates Foxg1-dependent progenitor cell expansion in the developing brain. Dev Cell. 2012. 22:871–878.
Article12. Eckey M, Kuphal S, Straub T, Rümmele P, Kremmer E, Bosserhoff AK, et al. Nucleosome remodeler SNF2L suppresses cell proliferation and migration and attenuates Wnt signaling. Mol Cell Biol. 2012. 32:2359–2371.
Article13. Xia Y, Jiang B, Zou Y, Gao G, Shang L, Chen B, et al. Sp1 and CREB regulate basal transcription of the human SNF2L gene. Biochem Biophys Res Commun. 2008. 368:438–444.
Article14. Lazzaro MA, Pépin D, Pescador N, Murphy BD, Vanderhyden BC, Picketts DJ. The imitation switch protein SNF2L regulates steroidogenic acute regulatory protein expression during terminal differentiation of ovarian granulosa cells. Mol Endocrinol. 2006. 20:2406–2417.
Article15. Conkright MD, Guzmán E, Flechner L, Su AI, Hogenesch JB, Montminy M. Genome-wide analysis of CREB target genes reveals a core promoter requirement for cAMP responsiveness. Mol Cell. 2003. 11:1101–1108.
Article16. Love TM, de Jesus R, Kean JA, Sheng Q, Leger A, Schaffhausen B. Activation of CREB/ATF sites by polyomavirus large T antigen. J Virol. 2005. 79:4180–4190.
Article17. Kehat I, Hasin T, Aronheim A. The role of basic leucine zipper protein-mediated transcription in physiological and pathological myocardial hypertrophy. Ann N Y Acad Sci. 2006. 1080:97–109.
Article18. Beck MR, DeKoster GT, Hambly DM, Gross ML, Cistola DP, Goldman WE. Structural features responsible for the biological stability of Histoplasma's virulence factor CBP. Biochemistry. 2008. 47:4427–4438.
Article19. DU C, Yang DM, Zhang PX, Deng L, Jiang BG. Neuronal differentiation of PC12 cells induced by sciatic nerve and optic nerve conditioned medium. Chin Med J (Engl). 2010. 123:351–355.20. Agholme L, Lindström T, Kågedal K, Marcusson J, Hallbeck M. An in vitro model for neuroscience: differentiation of SH-SY5Y cells into cells with morphological and biochemical characteristics of mature neurons. J Alzheimers Dis. 2010. 20:1069–1082.
Article21. Toulouse A, Collins GC, Sullivan AM. Neurotrophic effects of growth/differentiation factor 5 in a neuronal cell line. Neurotox Res. 2012. 21:256–265.
Article22. Dong L, Wang W, Wang F, Stoner M, Reed JC, Harigai M, et al. Mechanisms of transcriptional activation of bcl-2 gene expression by 17beta-estradiol in breast cancer cells. J Biol Chem. 1999. 274:32099–32107.
Article23. Atlas E, Stramwasser M, Mueller CR. A CREB site in the BRCA1 proximal promoter acts as a constitutive transcriptional element. Oncogene. 2001. 20:7110–7114.
Article24. Ogasawara K, Terada T, Asaka J, Katsura T, Inui K. Human organic anion transporter 3 gene is regulated constitutively and inducibly via a cAMP-response element. J Pharmacol Exp Ther. 2006. 319:317–322.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Molecular analysis of AQP2 promoter. I. cAMP-dependent regulation of mouse AQP2 gene
- Functional Characterization of cAMP-Regulated Gene, CAR1, in Cryptococcus neoformans
- Characterization of Cyclic AMP Response Element (CRE) in the Promoter of the Rat Thyrotropin Releasing Hormone (TRH) Gene
- Cell-type-specific Modulation of Nonhomologous End Joining of Gamma Ray-induced DNA Double-strand Breaks by cAMP Signaling in Human Cancer Cells
- Regulation of fibronectin gene expression by cyclic AMP and phorbol myristate acetate in HT-1080 human fibrosarcoma cells