J Korean Med Sci.
2013 Feb;28(2):237-246. 10.3346/jkms.2013.28.2.237.
Improved Therapeutic Effect against Leukemia by a Combination of the Histone Methyltransferase Inhibitor Chaetocin and the Histone Deacetylase Inhibitor Trichostatin A
- Affiliations
-
- 1Genome Research Center for Hematopoietic Diseases, Chonnam National University Hwasun Hospital, Hwasun, Korea. hjoonk@chonnam.ac.kr
- 2Department of Hematology-Oncology, Chonnam National University Hwasun Hospital, Hwasun, Korea.
- 3Department of Preventive Medicine, College of Medicine, Seonam University, Namwon, Korea.
- 4Environmental Health Center for Childhood Leukemia and Cancer, Chonnam National University Hwasun Hospital, Hwasun, Korea.
- KMID: 1429188
- DOI: http://doi.org/10.3346/jkms.2013.28.2.237
Abstract
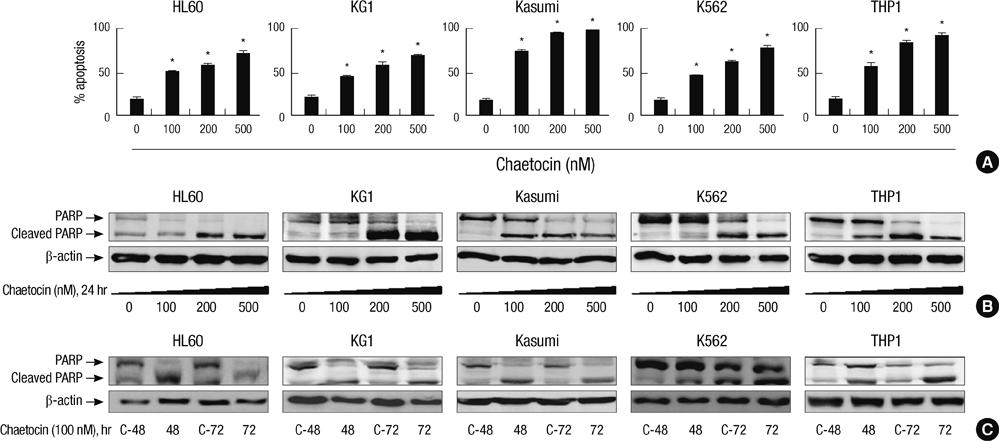
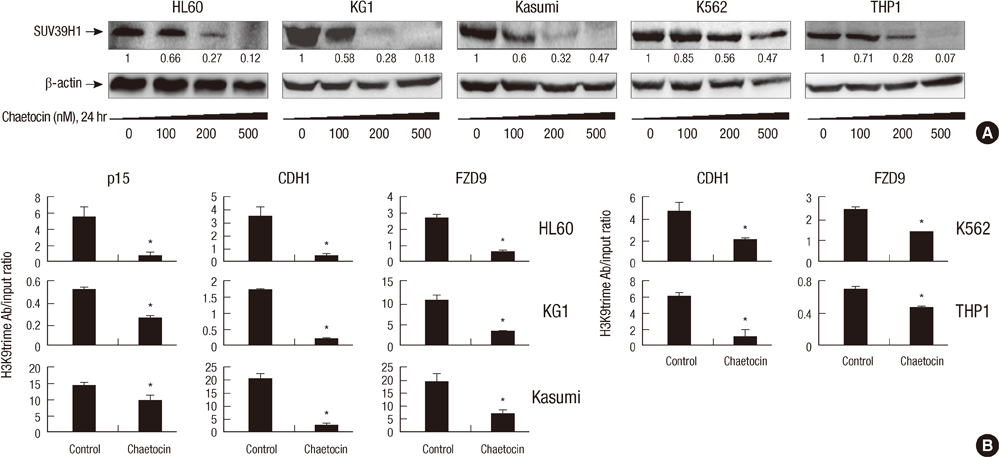
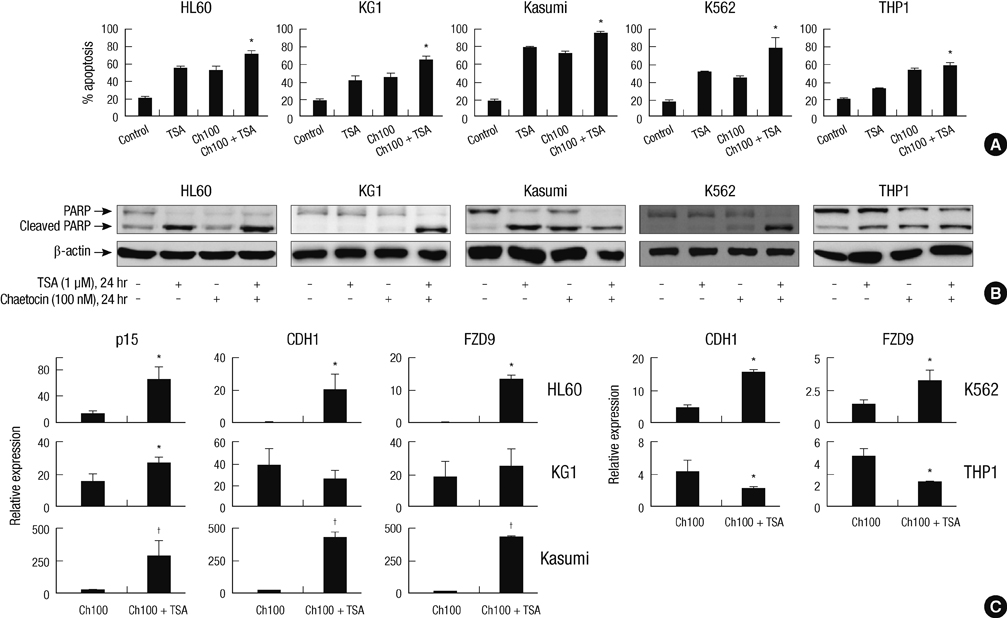
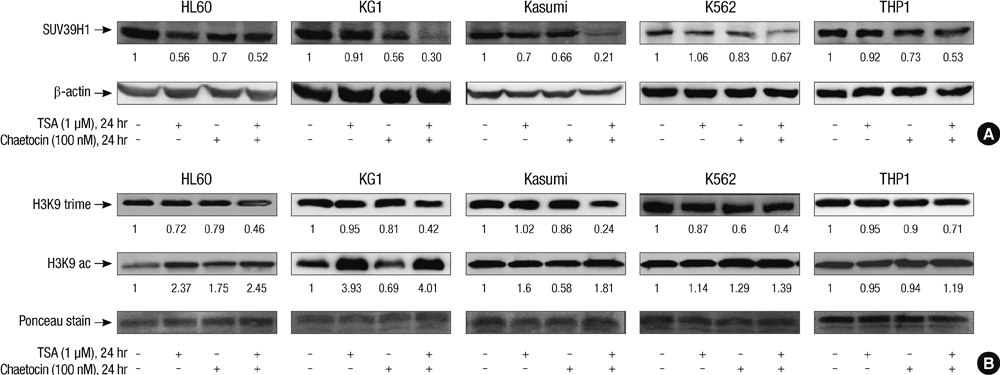
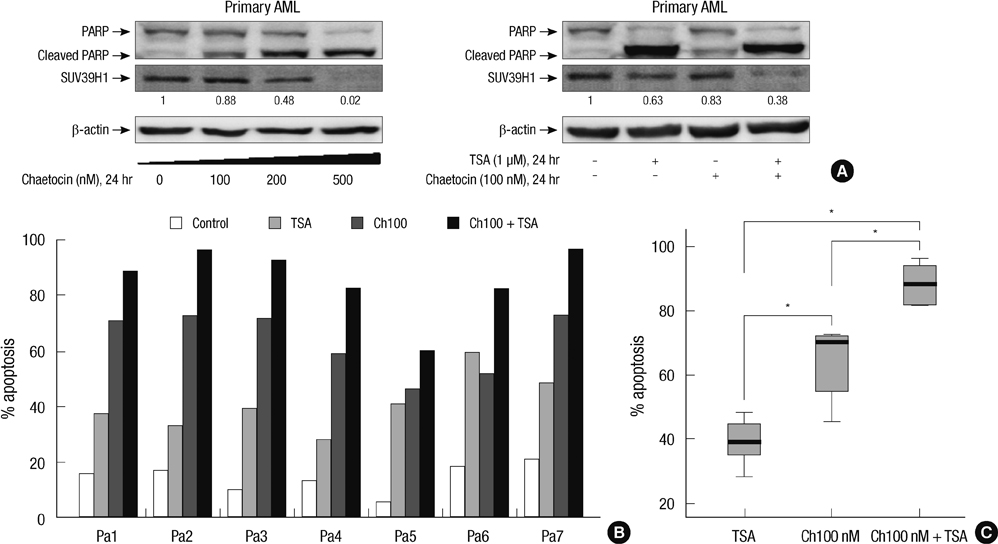
- SUV39H1 is a histone 3 lysine 9 (H3K9)-specific methyltransferase that is important for heterochromatin formation and the regulation of gene expression. Chaetocin specifically inhibits SUV39H1, resulted in H3K9 methylation reduction as well as reactivation of silenced genes in cancer cells. Histone deacetylase (HDAC) inhibitors inhibit deacetylases and accumulate high levels of acetylation lead to cell cycle arrest and apoptosis. In this study, we demonstrated that treatment with chaetocin enhanced apoptosis in human leukemia HL60, KG1, Kasumi, K562, and THP1 cells. In addition, chaetocin induced the expression of cyclin-dependent kinase inhibitor 2B (p15), E-cadherin (CDH1) and frizzled family receptor 9 (FZD9) through depletion of SUV39H1 and reduced H3K9 methylation in their promoters. Co-treatment with chaetocin and HDAC inhibitor trichostatin A (TSA) dramatically increased apoptosis and produced greater activation of genes. Furthermore, this combined treatment significantly increased loss of SUV39H1 and reduced histone H3K9 trimethylation responses accompanied by increased acetylation. Importantly, co-treatment with chaetocin and TSA produced potent antileukemic effects in leukemia cells derived from patients. These in vitro findings suggest that combination therapy with SUV39H1 and HDAC inhibitors may be of potential value in the treatment of leukemia.
MeSH Terms
-
Acetylation/drug effects
Adolescent
Adult
Aged
Apoptosis/*drug effects
Cadherins/metabolism
Cell Line, Tumor
Cyclin-Dependent Kinase Inhibitor p15/metabolism
DNA Methylation/drug effects
Enzyme Inhibitors/therapeutic use/*toxicity
Frizzled Receptors/metabolism
Gene Expression Regulation/drug effects
HL-60 Cells
Histone Deacetylase Inhibitors/therapeutic use/*toxicity
Histone-Lysine N-Methyltransferase/*antagonists & inhibitors/metabolism
Histones/genetics/metabolism
Humans
Hydroxamic Acids/therapeutic use/*toxicity
K562 Cells
Leukemia/drug therapy/metabolism/pathology
Leukemia, Myeloid, Acute/genetics/metabolism/pathology
Male
Middle Aged
Piperazines/therapeutic use/toxicity
Promoter Regions, Genetic
Young Adult
Cadherins
Cyclin-Dependent Kinase Inhibitor p15
Enzyme Inhibitors
Frizzled Receptors
Histone Deacetylase Inhibitors
Histones
Hydroxamic Acids
Piperazines
Histone-Lysine N-Methyltransferase
Figure
Reference
-
1. Chen J, Odenike O, Rowley JD. Leukaemogenesis: more than mutant genes. Nat Rev Cancer. 2010. 10:23–36.2. Krug U, Ganser A, Koeffler HP. Tumor suppressor genes in normal and malignant hematopoiesis. Oncogene. 2002. 21:3475–3495.3. Tran HT, Kim HN, Lee IK, Kim YK, Ahn JS, Yang DH, Lee JJ, Kim HJ. DNA methylation changes following 5-azacitidine treatment in patients with myelodysplastic syndrome. J Korean Med Sci. 2011. 26:207–213.4. Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004. 4:143–153.5. Lachner M, Jenuwein T. The many faces of histone lysine methylation. Curr Opin Cell Biol. 2002. 14:286–298.6. Vaute O, Nicolas E, Vandel L, Trouche D. Functional and physical interaction between the histone methyl transferase Suv39H1 and histone deacetylases. Nucleic Acids Res. 2002. 30:475–481.7. Fujita N, Watanabe S, Ichimura T, Tsuruzoe S, Shinkai Y, Tachibana M, Chiba T, Nakao M. Methyl-CpG binding domain 1 (MBD1) interacts with the Suv39h1-HP1 heterochromatic complex for DNA methylation-based transcriptional repression. J Biol Chem. 2003. 278:24132–24138.8. Braig M, Lee S, Loddenkemper C, Rudolph C, Peters AH, Schlegelberger B, Stein H, DÖrken B, Jenuwein T, Schmitt CA. Oncogene-induced senescence as an initial barrier in lymphoma development. Nature. 2005. 436:660–665.9. Kang MY, Lee BB, Kim YH, Chang DK, Park SK, Chun HK, Song SY, Park J, Kim DH. Association of the SUV39H1 histone methyltransferase with the DNA methyltransferase 1 at mRNA expression level in primary colorectal cancer. Int J Cancer. 2007. 121:2192–2197.10. Watanabe H, Soejima K, Yasuda H, Kawada I, Nakachi I, Yoda S, Naoki K, Ishizaka A. Deregulation of histone lysine methyltransferases contributes to oncogenic transformation of human bronchoepithelial cells. Cancer Cell Int. 2008. 8:15.11. Lakshmikuttyamma A, Scott SA, DeCoteau JF, Geyer CR. Reexpression of epigenetically silenced AML tumor suppressor genes by SUV39H1 inhibition. Oncogene. 2010. 29:576–588.12. Chaib H, Nebbioso A, Prebet T, Castellano R, Garbit S, Restouin A, Vey N, Altucci L, Collette Y. Anti-leukemia activity of chaetocin via death receptor-dependent apoptosis and dual modulation of the histone methyl-transferase SUV39H1. Leukemia. 2012. 26:662–674.13. Lukásová E, Koristek Z, Falk M, Kozubek S, Grigoryev S, Kozubek M, Ondrej V, Kroupová I. Methylation of histones in myeloid leukemias as a potential marker of granulocyte abnormalities. J Leukoc Biol. 2005. 77:100–111.14. Jiang Y, Dunbar A, Gondek LP, Mohan S, Rataul M, O'Keefe C, Sekeres M, Saunthararajah Y, Maciejewski JP. Aberrant DNA methylation is a dominant mechanism in MDS progression to AML. Blood. 2009. 113:1315–1325.15. Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3-9. Nat Chem Biol. 2005. 1:143–145.16. Tibodeau JD, Benson LM, Isham CR, Owen WG, Bible KC. The anticancer agent chaetocin is a competitive substrate and inhibitor of thioredoxin reductase. Antioxid Redox Signal. 2009. 11:1097–1106.17. Isham CR, Tibodeau JD, Bossou AR, Merchan JR, Bible KC. The anticancer effects of chaetocin are independent of programmed cell death and hypoxia, and are associated with inhibition of endothelial cell proliferation. Br J Cancer. 2012. 106:314–323.18. Isham CR, Tibodeau JD, Jin W, Xu R, Timm MM, Bible KC. Chaetocin: a promising new antimyeloma agent with in vitro and in vivo activity mediated via imposition of oxidative stress. Blood. 2007. 109:2579–2588.19. Fiskus W, Wang Y, Sreekumar A, Buckley KM, Shi H, Jillella A, Ustun C, Rao R, Fernandez P, Chen J, et al. Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood. 2009. 114:2733–2743.20. Fiskus W, Pranpat M, Balasis M, Herger B, Rao R, Chinnaiyan A, Atadja P, Bhalla K. Histone deacetylase inhibitors deplete enhancer of zeste 2 and associated polycomb repressive complex 2 proteins in human acute leukemia cells. Mol Cancer Ther. 2006. 5:3096–3104.21. Cherrier T, Suzanne S, Redel L, Calao M, Marban C, Samah B, Mukerjee R, Schwartz C, Gras G, Sawaya BE, et al. P21(WAF1) gene promoter is epigenetically silenced by CTIP2 and SUV39H1. Oncogene. 2009. 28:3380–3389.22. Spensberger D, Delwel R. A novel interaction between the proto-oncogene Evi1 and histone methyltransferases, SUV39H1 and G9a. FEBS Lett. 2008. 582:2761–2767.23. Chakraborty S, Sinha KK, Senyuk V, Nucifora G. SUV39H1 interacts with AML1 and abrogates AML1 transactivity: AML1 is methylated in vivo. Oncogene. 2003. 22:5229–5237.24. Kokura K, Sun L, Bedford MT, Fang J. Methyl-H3K9-binding protein MPP8 mediates E-cadherin gene silencing and promotes tumour cell motility and invasion. EMBO J. 2010. 29:3673–3687.25. Karasawa T, Yokokura H, Kitajewski J, Lombroso PJ. Frizzled-9 is activated by Wnt-2 and functions in Wnt/beta -catenin signaling. J Biol Chem. 2002. 277:37479–37486.26. Bots M, Johnstone RW. Rational combinations using HDAC inhibitors. Clin Cancer Res. 2009. 15:3970–3977.27. Kosugi H, Towatari M, Hatano S, Kitamura K, Kiyoi H, Kinoshita T, Tanimoto M, Murate T, Kawashima K, Saito H, et al. Histone deacetylase inhibitors are the potent inducer/enhancer of differentiation in acute myeloid leukemia: a new approach to anti-leukemia therapy. Leukemia. 1999. 13:1316–1324.28. Yang G, Thompson MA, Brandt SJ, Hiebert SW. Histone deacetylase inhibitors induce the degradation of the t(8;21) fusion oncoprotein. Oncogene. 2007. 26:91–101.29. Pabst T, Mueller BU, Harakawa N, Schoch C, Haferlach T, Behre G, Hiddemann W, Zhang DE, Tenen DG. AML1-ETO downregulates the granulocytic differentiation factor C/EBPalpha in t(8;21) myeloid leukemia. Nat Med. 2001. 7:444–451.30. Serrano M, Lee H, Chin L, Cordon-Cardo C, Beach D, DePinho RA. Role of the INK4a locus in tumor suppression and cell mortality. Cell. 1996. 85:27–37.31. Karayiannakis AJ, Syrigos KN, Chatzigianni E, Papanikolaou S, Alexiou D, Kalahanis N, Rosenberg T, Bastounis E. Aberrant E-cadherin expression associated with loss of differentiation and advanced stage in human pancreatic cancer. Anticancer Res. 1998. 18:4177–4180.32. Sulzer MA, Leers MP, van Noord JA, Bollen EC, Theunissen PH. Reduced E-cadherin expression is associated with increased lymph node metastasis and unfavorable prognosis in non-small cell lung cancer. Am J Respir Crit Care Med. 1998. 157:1319–1323.33. Khan NI, Bradstock KF, Bendall LJ. Activation of Wnt/beta-catenin pathway mediates growth and survival in B-cell progenitor acute lymphoblastic leukaemia. Br J Haematol. 2007. 138:338–348.34. Habold C, Poehlmann A, Bajbouj K, Hartig R, Korkmaz KS, Roessner A, Schneider-Stock R. Trichostatin A causes p53 to switch oxidative-damaged colorectal cancer cells from cell cycle arrest into apoptosis. J Cell Mol Med. 2008. 12:607–621.35. Muscolini M, Cianfrocca R, Sajeva A, Mozzetti S, Ferrandina G, Costanzo A, Tuosto L. Trichostatin A up-regulates p73 and induces Bax-dependent apoptosis in cisplatin-resistant ovarian cancer cells. Mol Cancer Ther. 2008. 7:1410–1419.36. Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006. 6:38–51.37. Zhang Y, Yu G, Wang D, Hu Y, Lei W. ERK1/2 activation plays important roles in the opposite effects of Trichostatin A in non-cancer and cancer cells. Toxicon. 2011. 57:932–937.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Apoptotic Effect of Combination Treatment with a Proteasome Inhibitor, Lactacystin, and a Histone Deacetylase Inhibitor, Trichostatin A, on MCF-7 Cells
- Activation of ATM-dependent DNA Damage Signal Pathway by a Histone Deacetylase Inhibitor, Trichostatin A
- Trichostatin A, a Histone Deacetylase Inhibitor, Potentiated Cytotoxic Effect of Ionizing Radiation in Human Head and Neck Cancer Cell Lines
- Role of histone deacetylase activity in the developing lateral line neuromast of zebrafish larvae
- Recovery of Genes Epigenetically Altered by the Histone Deacetylase Inhibitor Scriptaid and Demethylating Agent 5-Azacytidine in Human Leukemia Cells