Ann Dermatol.
2020 Apr;32(2):122-129. 10.5021/ad.2020.32.2.122.
Epigenetic Regulation of Filaggrin Gene Expression in Human Epidermal Keratinocytes
- Affiliations
-
- 1Bio-Integration Research Center for Nutra-Pharmaceutical Epigenetics, Chung-Ang University, Seoul, Korea. uromyung@cau.ac.kr
- 2Department of Urology, Chung-Ang University College of Medicine, Seoul, Korea.
- 3Department of Dermatology, Chung-Ang University College of Medicine, Seoul, Korea.
- KMID: 2471338
- DOI: http://doi.org/10.5021/ad.2020.32.2.122
Abstract
- BACKGROUND
Loss-of-function mutations in the filaggrin gene (FLG), which encodes an epidermal protein crucial for the formation of a functional skin barrier, have been identified as a major predisposing factor in the etiopathogenesis of atopic dermatitis (AD). Recent reports of relatively low frequencies of FLG-null mutations among specific ethnic groups with AD necessitated analysis of the epigenetic regulation which may control FLG expression without altering its DNA sequence.
OBJECTIVE
The study aimed to identify DNA methylation-dependent regulation of FLG expression.
METHODS
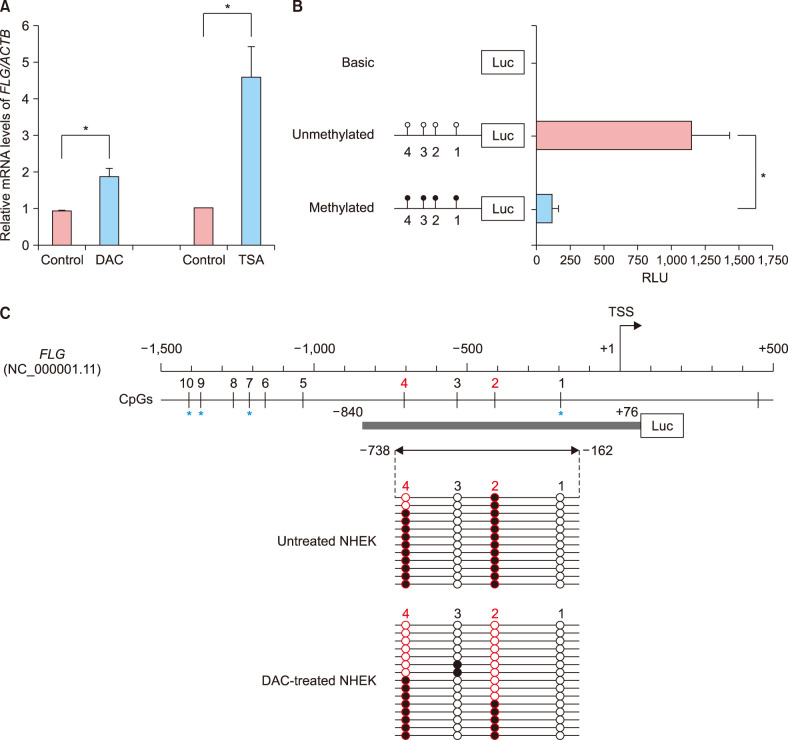
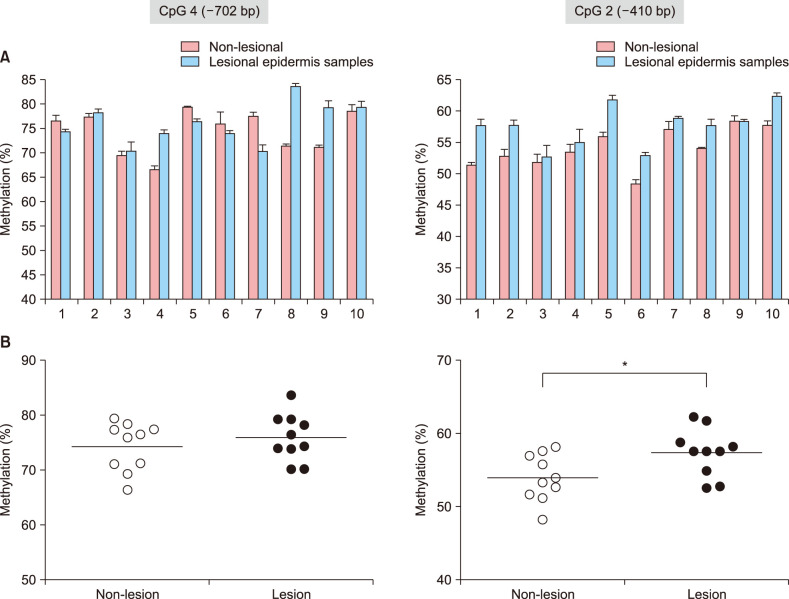
Quantitative polymerase chain reaction was performed to determine the restoration of FLG mRNA expression in normal human epidermal keratinocyte (NHEK) cells after treatment with epigenetic modulating agents. Bisulfite genomic sequencing and pyrosequencing analyses of the FLG promoter region were conducted to identify the citical CpG sites relevant to FLG expression. We performed small-scale pilot study for epidermal tissues obtained from Korean patients with severe AD.
RESULTS
We here show that DNA methylation in the FLG with non-CpG island promoter is responsible for the transcriptional regulation of FLG in undifferentiated NHEK cells. The methylation frequencies in a single CpG site of the FLG promoter were significantly higher in lesional epidermis than those in matched nonlesional epidermis of subjects with severe AD.
CONCLUSION
Our in vitro and clinical studies point to this unique CpG site as a potential DNA methylation marker of FLG, which can be a promising therapeutic target in the complications of filaggrin-related skin barrier dysfunction as well as in AD.
Keyword
MeSH Terms
Figure
Reference
-
1. Brown SJ, McLean WH. One remarkable molecule: filaggrin. J Invest Dermatol. 2012; 132(3 Pt 2):751–762. PMID: 22158554.
Article2. Elias PM, Steinhoff M. “Outside-to-inside” (and now back to “outside”) pathogenic mechanisms in atopic dermatitis. J Invest Dermatol. 2008; 128:1067–1070. PMID: 18408746.
Article3. Smith FJ, Irvine AD, Terron-Kwiatkowski A, Sandilands A, Campbell LE, Zhao Y, et al. Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet. 2006; 38:337–342. PMID: 16444271.
Article4. Palmer CN, Irvine AD, Terron-Kwiatkowski A, Zhao Y, Liao H, Lee SP, et al. Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet. 2006; 38:441–446. PMID: 16550169.5. Sandilands A, Terron-Kwiatkowski A, Hull PR, O'Regan GM, Clayton TH, Watson RM, et al. Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet. 2007; 39:650–654. PMID: 17417636.6. Nomura T, Sandilands A, Akiyama M, Liao H, Evans AT, Sakai K, et al. Unique mutations in the filaggrin gene in Japanese patients with ichthyosis vulgaris and atopic dermatitis. J Allergy Clin Immunol. 2007; 119:434–440. PMID: 17291859.7. Hamada T, Sandilands A, Fukuda S, Sakaguchi S, Ohyama B, Yasumoto S, et al. De novo occurrence of the filaggrin mutation p.R501X with prevalent mutation c.3321delA in a Japanese family with ichthyosis vulgaris complicated by atopic dermatitis. J Invest Dermatol. 2008; 128:1323–1325. PMID: 18007582.
Article8. Zhang X, Liu S, Chen X, Zhou B, Liu D, Lei G, et al. Novel and recurrent mutations in the filaggrin gene in Chinese patients with ichthyosis vulgaris. Br J Dermatol. 2010; 163:63–69. PMID: 20222934.
Article9. Zhang H, Guo Y, Wang W, Shi M, Chen X, Yao Z. Mutations in the filaggrin gene in Han Chinese patients with atopic dermatitis. Allergy. 2011; 66:420–427. PMID: 21039602.
Article10. Chen H, Common JE, Haines RL, Balakrishnan A, Brown SJ, Goh CS, et al. Wide spectrum of filaggrin-null mutations in atopic dermatitis highlights differences between Singaporean Chinese and European populations. Br J Dermatol. 2011; 165:106–114. PMID: 21428977.
Article11. Hsu CK, Akiyama M, Nemoto-Hasebe I, Nomura T, Sandilands A, Chao SC, et al. Analysis of Taiwanese ichthyosis vulgaris families further demonstrates differences in FLG mutations between European and Asian populations. Br J Dermatol. 2009; 161:448–451. PMID: 19416262.12. Kim EJ, Jeong MS, Li K, Park MK, Lee MK, Yoon Y, et al. Genetic polymorphism of FLG in Korean ichthyosis vulgaris patients. Ann Dermatol. 2011; 23:170–176. PMID: 21747615.13. Park J, Jekarl DW, Kim Y, Kim J, Kim M, Park YM. Novel FLG null mutations in Korean patients with atopic dermatitis and comparison of the mutational spectra in Asian populations. J Dermatol. 2015; 42:867–873. PMID: 25997159.14. Winge MC, Bilcha KD, Liedén A, Shibeshi D, Sandilands A, Wahlgren CF, et al. Novel filaggrin mutation but no other loss-of-function variants found in Ethiopian patients with atopic dermatitis. Br J Dermatol. 2011; 165:1074–1080. PMID: 21692775.
Article15. Margolis DJ, Gupta J, Apter AJ, Hoffstad O, Papadopoulos M, Rebbeck TR, et al. Exome sequencing of filaggrin and related genes in African-American children with atopic dermatitis. J Invest Dermatol. 2014; 134:2272–2274. PMID: 24608987.
Article16. Polcari I, Becker L, Stein SL, Smith MS, Paller AS. Filaggrin gene mutations in African Americans with both ichthyosis vulgaris and atopic dermatitis. Pediatr Dermatol. 2014; 31:489–492. PMID: 24920311.
Article17. Thawer-Esmail F, Jakasa I, Todd G, Wen Y, Brown SJ, Kroboth K, et al. South African amaXhosa patients with atopic dermatitis have decreased levels of filaggrin breakdown products but no loss-of-function mutations in filaggrin. J Allergy Clin Immunol. 2014; 133:280–282. 282.e1–282.e2. PMID: 24369804.
Article18. Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009; 23:781–783. PMID: 19339683.19. Jaenisch R, Bird A. Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet. 2003; 33:245–254. PMID: 12610534.
Article20. Botchkarev VA, Gdula MR, Mardaryev AN, Sharov AA, Fessing MY. Epigenetic regulation of gene expression in keratinocytes. J Invest Dermatol. 2012; 132:2505–2521. PMID: 22763788.
Article21. Schübeler D. Function and information content of DNA methylation. Nature. 2015; 517:321–326. PMID: 25592537.
Article22. Jones PA. Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet. 2012; 13:484–492. PMID: 22641018.
Article23. Li Y, Sawalha AH, Lu Q. Aberrant DNA methylation in skin diseases. J Dermatol Sci. 2009; 54:143–149. PMID: 19395242.
Article24. Ziyab AH, Karmaus W, Holloway JW, Zhang H, Ewart S, Arshad SH. DNA methylation of the filaggrin gene adds to the risk of eczema associated with loss-of-function variants. J Eur Acad Dermatol Venereol. 2013; 27:e420–e423. PMID: 23003573.
Article25. Lee J, Han JH, Jang A, Kim JW, Hong SA, Myung SC. DNA methylation-mediated downregulation of DEFB1 in prostate cancer cells. PLoS One. 2016; 11:e0166664. PMID: 27835705.
Article26. Alaskhar Alhamwe B, Khalaila R, Wolf J, von Bülow V, Harb H, Alhamdan F, et al. Histone modifications and their role in epigenetics of atopy and allergic diseases. Allergy Asthma Clin Immunol. 2018; 14:39. PMID: 29796022.
Article27. Bhardwaj N. MicroRNAs in atopic dermatitis: a review. J Transl Genet Genom. 2017; 1:15–22.
Article28. Gardiner-Garden M, Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987; 196:261–282. PMID: 3656447.
Article29. Stadler MB, Murr R, Burger L, Ivanek R, Lienert F, Schöler A, et al. DNA-binding factors shape the mouse methylome at distal regulatory regions. Nature. 2011; 480:490–495. PMID: 22170606.
Article30. Ziller MJ, Gu H, Müller F, Donaghey J, Tsai LT, Kohlbacher O, et al. Charting a dynamic DNA methylation landscape of the human genome. Nature. 2013; 500:477–481. PMID: 23925113.
Article31. Rodríguez E, Baurecht H, Wahn AF, Kretschmer A, Hotze M, Zeilinger S, et al. An integrated epigenetic and transcriptomic analysis reveals distinct tissue-specific patterns of DNA methylation associated with atopic dermatitis. J Invest Dermatol. 2014; 134:1873–1883. PMID: 24739813.
Article32. Tan HT, Ellis JA, Koplin JJ, Martino D, Dang TD, Suaini N, et al. HealthNuts Study Investigators. Methylation of the filaggrin gene promoter does not affect gene expression and allergy. Pediatr Allergy Immunol. 2014; 25:608–610. PMID: 24912553.
Article33. Weber M, Hellmann I, Stadler MB, Ramos L, Pääbo S, Rebhan M, et al. Distribution, silencing potential and evolutionary impact of promoter DNA methylation in the human genome. Nat Genet. 2007; 39:457–466. PMID: 17334365.
Article34. Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008; 454:766–770. PMID: 18600261.
Article35. Kezic S, O'Regan GM, Yau N, Sandilands A, Chen H, Campbell LE, et al. Levels of filaggrin degradation products are influenced by both filaggrin genotype and atopic dermatitis severity. Allergy. 2011; 66:934–940. PMID: 21261659.
Article36. Bataille V, Lens M, Spector TD. The use of the twin model to investigate the genetics and epigenetics of skin diseases with genomic, transcriptomic and methylation data. J Eur Acad Dermatol Venereol. 2012; 26:1067–1073. PMID: 22243446.
Article37. Tost J, Dunker J, Gut IG. Analysis and quantification of multiple methylation variable positions in CpG islands by Pyrosequencing. Biotechniques. 2003; 35:152–156. PMID: 12866415.
Article38. Rakyan VK, Down TA, Balding DJ, Beck S. Epigenome-wide association studies for common human diseases. Nat Rev Genet. 2011; 12:529–541. PMID: 21747404.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Adiponectin Upregulates Filaggrin Expression via SIRT1-Mediated Signaling in Human Normal Keratinocytes
- Epigenetic Modulation of Gene Expression during Keratinocyte Differentiation
- Immunohistochemical Study of Expression of Involucrin, Loricrin, Filaggrin and Bcl-2 in Psoriasis
- Epigenetic Regulation of Cytokine Gene Expression in T Lymphocytes
- HOTAIR Long Non-coding RNA: Characterizing the Locus Features by the In Silico Approaches