Allergy Asthma Immunol Res.
2020 Jan;12(1):137-148. 10.4168/aair.2020.12.1.137.
Antibiotics-Induced Dysbiosis of Intestinal Microbiota Aggravates Atopic Dermatitis in Mice by Altered Short-Chain Fatty Acids
- Affiliations
-
- 1Department of Internal Medicine, College of Veterinary Medicine, Chonnam National University, Gwangju, Korea.
- 2Asan Institute for Life Sciences, University of Ulsan College of Medicine, Seoul, Korea.
- 3Department of Pediatrics, Childhood Asthma Atopy Center, Asan Medical Center, University of Ulsan College of Medicine, Seoul, Korea. sjhong@amc.seoul.kr
- 4Environmental Health Center, Asan Medical Center, Seoul, Korea.
- KMID: 2462578
- DOI: http://doi.org/10.4168/aair.2020.12.1.137
Abstract
- PURPOSE
Alterations in the intestinal microbiota in early life affects the development of atopic dermatitis (AD) in humans. This study aimed to further investigate the effects of gut dysbiosis in early life in an ovalbumin (OVA)-induced mouse model of AD.
METHODS
The AD mouse model was developed by serial OVA sensitization and mice were treated with an antibiotic cocktail in their drinking water for 2 weeks before primary sensitization. Probiotics (Lactobacillus rhamnosus, 1 × 10â¹ CFU) or 100 µL of fresh fecal supernatant were orally administered daily from 1 week before the ï¬rst sensitization until the end of the study.
RESULTS
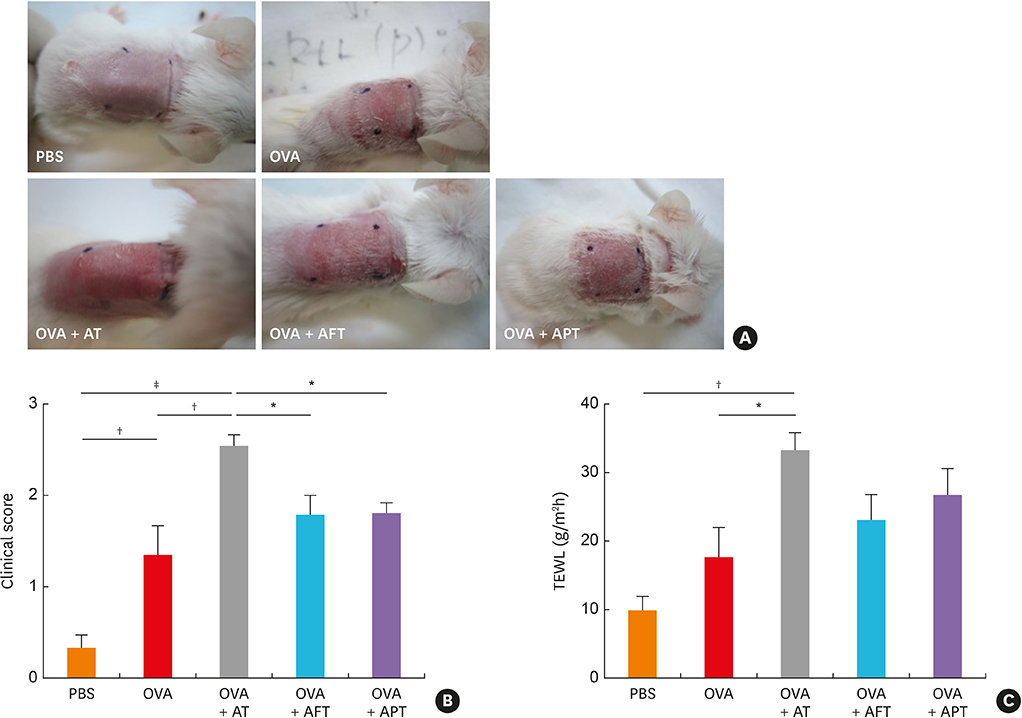
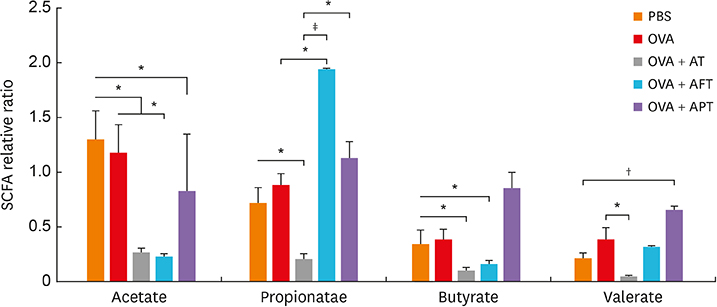
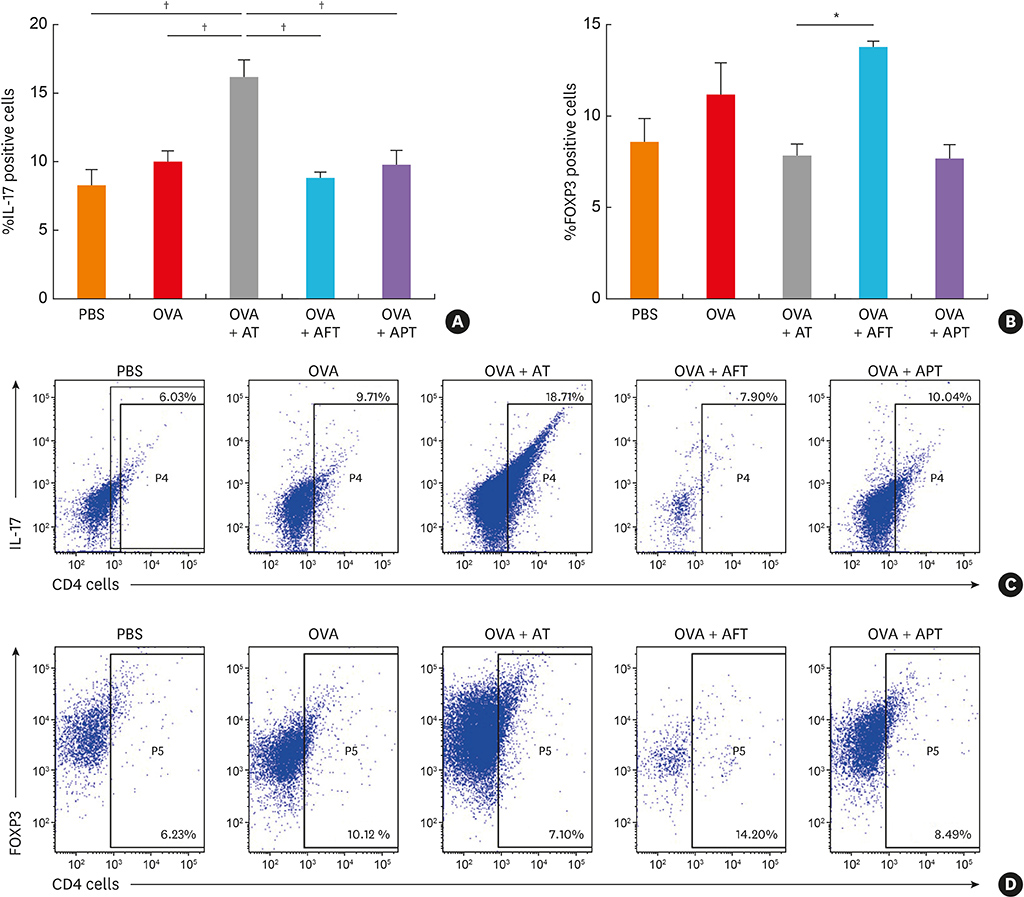
The AD mice which received antibiotics had significantly aggravated phenotypes, including clinical score, transepidermal water loss, and histopathology, compared to those treated with healthy feces or probiotics. Total systemic immunoglobulin E production and skin interleukin (IL) 4 levels were significantly increased in the antibiotic-treated mice compared to the other groups. Antibiotic treatment also increased the levels of IL17 and group 3 innate lymphoid cells (ILC3) in the gut and significantly suppressed the production of short-chain fatty acids (SCFAs) and decreased the number FOXP3⺠cells.
CONCLUSIONS
Our results suggest that the status of the gut microbiota in early life in the mouse may play a crucial role in AD development through intestinal SCFA production through regulate the numbers of CD4âºIL17âº/CD4âºFOXP3⺠regulatory T cells and ILC3s.
Keyword
MeSH Terms
-
Animals
Anti-Bacterial Agents
Cytokines
Dermatitis, Atopic*
Drinking Water
Dysbiosis*
Fatty Acids
Fatty Acids, Volatile*
Feces
Gastrointestinal Microbiome*
Humans
Immunoglobulin E
Immunoglobulins
Interleukins
Intestines
Lymphocytes
Mice*
Microbiota
Ovalbumin
Ovum
Phenotype
Probiotics
Skin
T-Lymphocytes, Regulatory
Water
Anti-Bacterial Agents
Cytokines
Drinking Water
Fatty Acids
Fatty Acids, Volatile
Immunoglobulin E
Immunoglobulins
Interleukins
Ovalbumin
Water
Figure
Reference
-
1. Okada H, Kuhn C, Feillet H, Bach JF. The ‘hygiene hypothesis’ for autoimmune and allergic diseases: an update. Clin Exp Immunol. 2010; 160:1–9.
Article2. Lee SY, Kwon JW, Seo JH, Song YH, Kim BJ, Yu J, et al. Prevalence of atopy and allergic diseases in Korean children: associations with a farming environment and rural lifestyle. Int Arch Allergy Immunol. 2012; 158:168–174.
Article3. Alduraywish SA, Lodge CJ, Campbell B, Allen KJ, Erbas B, Lowe AJ, et al. The march from early life food sensitization to allergic disease: a systematic review and meta-analyses of birth cohort studies. Allergy. 2016; 71:77–89.
Article4. Lee JY, Seo JH, Kwon JW, Yu J, Kim BJ, Lee SY, et al. Exposure to gene-environment interactions before 1 year of age may favor the development of atopic dermatitis. Int Arch Allergy Immunol. 2012; 157:363–371.
Article5. Cahenzli J, Köller Y, Wyss M, Geuking MB, McCoy KD. Intestinal microbial diversity during early-life colonization shapes long-term IgE levels. Cell Host Microbe. 2013; 14:559–570.
Article6. Fujimura KE, Lynch SV. Microbiota in allergy and asthma and the emerging relationship with the gut microbiome. Cell Host Microbe. 2015; 17:592–602.
Article7. Simonyte Sjödin K, Vidman L, Rydén P, West CE. Emerging evidence of the role of gut microbiota in the development of allergic diseases. Curr Opin Allergy Clin Immunol. 2016; 16:390–395.
Article8. West CE, Renz H, Jenmalm MC, Kozyrskyj AL, Allen KJ, Vuillermin P, et al. The gut microbiota and inflammatory noncommunicable diseases: associations and potentials for gut microbiota therapies. J Allergy Clin Immunol. 2015; 135:3–13.
Article9. Lee SY, Yu J, Ahn KM, Kim KW, Shin YH, Lee KS, et al. Additive effect between IL-13 polymorphism and cesarean section delivery/prenatal antibiotics use on atopic dermatitis: a birth cohort study (COCOA). PLoS One. 2014; 9:e96603.
Article10. Kim HJ, Kim YJ, Lee SH, Yu J, Jeong SK, Hong SJ. Effects of Lactobacillus rhamnosus on allergic march model by suppressing Th2, Th17, and TSLP responses via CD4(+)CD25(+)Foxp3(+) Tregs. Clin Immunol. 2014; 153:178–186.11. Kim HJ, Kim YJ, Kang MJ, Seo JH, Kim HY, Jeong SK, et al. A novel mouse model of atopic dermatitis with epicutaneous allergen sensitization and the effect of Lactobacillus rhamnosus . Exp Dermatol. 2012; 21:672–675.12. Song H, Yoo Y, Hwang J, Na YC, Kim HS. Faecalibacterium prausnitzii subspecies-level dysbiosis in the human gut microbiome underlying atopic dermatitis. J Allergy Clin Immunol. 2016; 137:852–860.13. Zeissig S, Blumberg RS. Life at the beginning: perturbation of the microbiota by antibiotics in early life and its role in health and disease. Nat Immunol. 2014; 15:307–310.
Article14. Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013; 504:446–450.
Article15. Smith MI, Yatsunenko T, Manary MJ, Trehan I, Mkakosya R, Cheng J, et al. Gut microbiomes of Malawian twin pairs discordant for kwashiorkor. Science. 2013; 339:548–554.
Article16. Kabat AM, Srinivasan N, Maloy KJ. Modulation of immune development and function by intestinal microbiota. Trends Immunol. 2014; 35:507–517.
Article17. Lee SY, Lee E, Park YM, Hong SJ. Microbiome in the gut-skin axis in atopic dermatitis. Allergy Asthma Immunol Res. 2018; 10:354–362.
Article18. Sharma G, Im SH. Probiotics as a potential immunomodulating pharmabiotics in allergic diseases: current status and future prospects. Allergy Asthma Immunol Res. 2018; 10:575–590.
Article19. Zanvit P, Konkel JE, Jiao X, Kasagi S, Zhang D, Wu R, et al. Antibiotics in neonatal life increase murine susceptibility to experimental psoriasis. Nat Commun. 2015; 6:8424.
Article20. Strzępa A, Majewska-Szczepanik M, Kowalczyk P, Woźniak D, Motyl S, Szczepanik M. Oral treatment with enrofloxacin early in life promotes Th2-mediated immune response in mice. Pharmacol Rep. 2016; 68:44–50.
Article21. Strzępa A, Majewska-Szczepanik M, Lobo FM, Wen L, Szczepanik M. Broad spectrum antibiotic enrofloxacin modulates contact sensitivity through gut microbiota in a murine model. J Allergy Clin Immunol. 2017; 140:121–133.e3.
Article22. Candela M, Rampelli S, Turroni S, Severgnini M, Consolandi C, De Bellis G, et al. Unbalance of intestinal microbiota in atopic children. BMC Microbiol. 2012; 12:95.
Article23. Abrahamsson TR, Jakobsson HE, Andersson AF, Björkstén B, Engstrand L, Jenmalm MC. Low diversity of the gut microbiota in infants with atopic eczema. J Allergy Clin Immunol. 2012; 129:434–440.
Article24. Fyhrquist N, Lehtimäki S, Lahl K, Savinko T, Lappeteläinen AM, Sparwasser T, et al. Foxp3+ cells control Th2 responses in a murine model of atopic dermatitis. J Invest Dermatol. 2012; 132:1672–1680.
Article25. Heimesaat MM, Bereswill S, Fischer A, Fuchs D, Struck D, Niebergall J, et al. Gram-negative bacteria aggravate murine small intestinal Th1-type immunopathology following oral infection with Toxoplasma gondii. J Immunol. 2006; 177:8785–8795.26. Chen X, Katchar K, Goldsmith JD, Nanthakumar N, Cheknis A, Gerding DN, et al. A mouse model of Clostridium difficile-associated disease. Gastroenterology. 2008; 135:1984–1992.27. Lefrancois L. Phenotypic complexity of intraepithelial lymphocytes of the small intestine. J Immunol. 1991; 147:1746–1751.28. Smith PM, Howitt MR, Panikov N, Michaud M, Gallini CA, Bohlooly-Y M, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013; 341:569–573.
Article29. Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013; 504:451–455.
Article30. Gevers D, Kugathasan S, Denson LA, Vázquez-Baeza Y, Van Treuren W, Ren B, et al. The treatment-naive microbiome in new-onset Crohn's disease. Cell Host Microbe. 2014; 15:382–392.
Article31. Cani PD, Bibiloni R, Knauf C, Waget A, Neyrinck AM, Delzenne NM, et al. Changes in gut microbiota control metabolic endotoxemia-induced inflammation in high-fat diet-induced obesity and diabetes in mice. Diabetes. 2008; 57:1470–1481.
Article32. Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, et al. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science. 2010; 328:228–231.
Article33. Chang SH, Dong C. Signaling of interleukin-17 family cytokines in immunity and inflammation. Cell Signal. 2011; 23:1069–1075.
Article34. Ouyang W, Kolls JK, Zheng Y. The biological functions of T helper 17 cell effector cytokines in inflammation. Immunity. 2008; 28:454–467.
Article35. Dubin PJ, Kolls JK. Th17 cytokines and mucosal immunity. Immunol Rev. 2008; 226:160–171.
Article36. Mortha A, Chudnovskiy A, Hashimoto D, Bogunovic M, Spencer SP, Belkaid Y, et al. Microbiota-dependent crosstalk between macrophages and ILC3 promotes intestinal homeostasis. Science. 2014; 343:1249288.
Article37. Bessman NJ, Sonnenberg GF. Emerging roles for antigen presentation in establishing host-microbiome symbiosis. Immunol Rev. 2016; 272:139–150.
Article38. Montaldo E, Juelke K, Romagnani C. Group 3 innate lymphoid cells (ILC3s): origin, differentiation, and plasticity in humans and mice. Eur J Immunol. 2015; 45:2171–2182.
Article39. Christensen GJ, Brüggemann H. Bacterial skin commensals and their role as host guardians. Benef Microbes. 2014; 5:201–215.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- FFA2 Activation Ameliorates 2,4-Dinitrochlorobenzene-Induced Atopic Dermatitis in Mice
- The Effects of Short-Chain Fatty Acids in Urological Diseases
- Regulation of Allergic Immune Responses by Microbial Metabolites
- The Role of Short Chain Fatty Acids in Irritable Bowel Syndrome
- High-fat-diet-modulated Gut Microbiota Promotes Intestinal Carcinogenesis