J Gastric Cancer.
2019 Sep;19(3):301-314. 10.5230/jgc.2019.19.e27.
Exosomal miR-181b-5p Downregulation in Ascites Serves as a Potential Diagnostic Biomarker for Gastric Cancer-associated Malignant Ascites
- Affiliations
-
- 1Department of Pharmaceutical Engineering, College of Science Engineering, Cheongju University, Cheongju, Korea.
- 2College of Pharmacy, Chungbuk National University, Cheongju, Korea.
- 3Department of Internal Medicine, Gyeongsang National University School of Medicine, Jinju, Korea.
- 4Department of Internal Medicine, Chonnam National University Hwasun Hospital, Hwasun, Korea.
- 5Department of Internal Medicine, Chonnam National University Medical School, Gwangju, Korea.
- 6Department of Internal Medicine, Chonbuk National University Medical School, Jeonju, Korea.
- 7Research Institute of Clinical Medicine of Chonbuk National University, Biomedical Research Institute of Chonbuk National University, Jeonju, Korea.
- 8Department Pathology, Chungbuk National University Hospital, Cheongju, Korea.
- 9Department of Pathology, Chungbuk National University College of Medicine, Cheongju, Korea.
- 10Department of Internal Medicine, Chungbuk National University Hospital, Cheongju, Korea. sook3529@hanmail.net
- 11Department of Internal Medicine, Chungbuk National University College of Medicine, Korea.
- KMID: 2458830
- DOI: http://doi.org/10.5230/jgc.2019.19.e27
Abstract
- PURPOSE
Peritoneal carcinomatosis in gastric cancer (GC) patients results in extremely poor prognosis. Malignant ascites samples are the most appropriate biological material to use to evaluate biomarkers for peritoneal carcinomatosis. This study identified exosomal MicroRNAs (miRNAs) differently expressed between benign liver cirrhosis-associated ascites (LC-ascites) and malignant gastric cancer-associated ascites (GC-ascites), and validated their role as diagnostic biomarkers for GC-ascites.
MATERIALS AND METHODS
Total RNA was extracted from exosomes isolated from 165 ascites samples (73 LC-ascites and 92 GC-ascites). Initially, microarrays were used to screen the expression levels of 2,006 miRNAs in the discovery cohort (n=22). Subsequently, quantitative reverse transcriptase-polymerase chain reaction (qRT-PCR) analyses were performed to validate the expression levels of selected exosomal miRNAs in the training (n=70) and validation (n=73) cohorts. Furthermore, carcinoembryonic antigen (CEA) levels were determined in ascites samples.
RESULTS
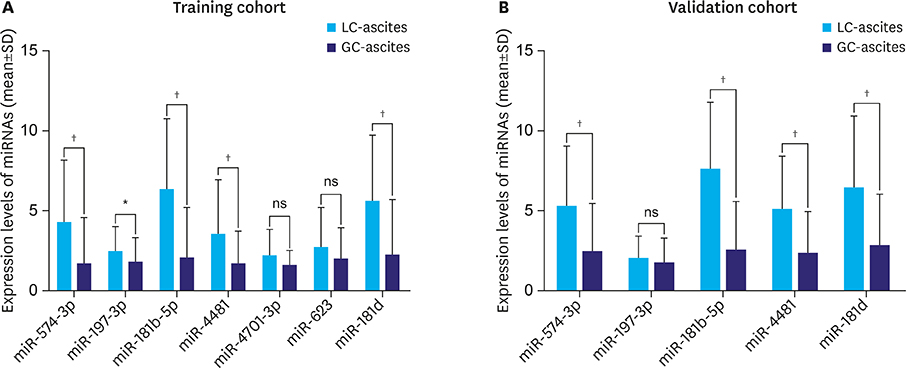
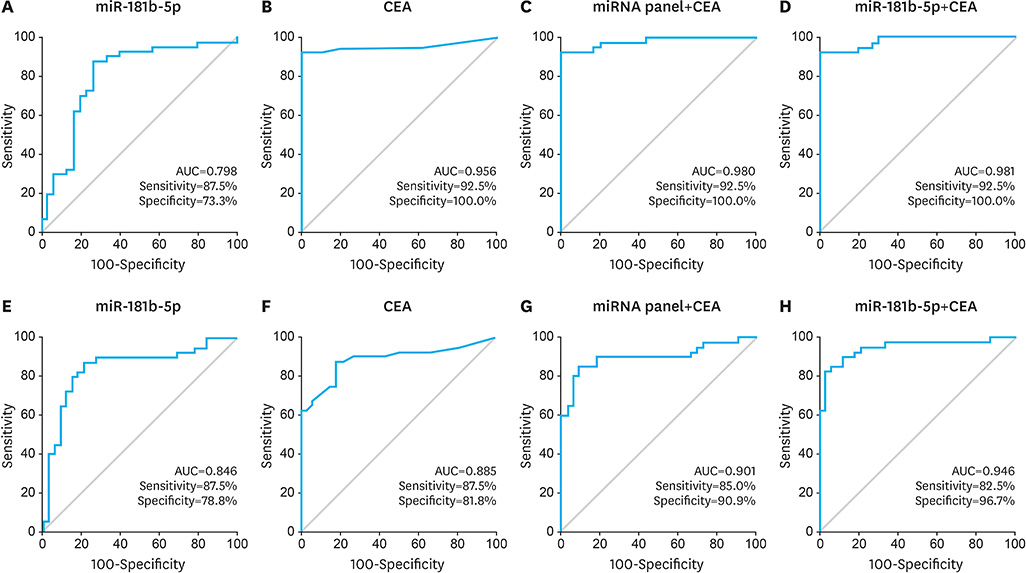
The miR-574-3p, miR-181b-5p, miR-4481, and miR-181d were significantly downregulated in the GC-ascites samples compared to the LC-ascites samples, and miR-181b-5p showed the best diagnostic performance for GC-ascites (area under the curve [AUC]=0.798 and 0.846 for the training and validation cohorts, respectively). The diagnostic performance of CEA for GC-ascites was improved by the combined analysis of miR-181b-5p and CEA (AUC=0.981 and 0.946 for the training and validation cohorts, respectively).
CONCLUSIONS
We identified exosomal miRNAs capable of distinguishing between non-malignant and GC-ascites, showing that the combined use of miR-181b-5p and CEA could improve diagnosis.
Keyword
MeSH Terms
Figure
Reference
-
1. Leiting JL, Grotz TE. Optimizing outcomes for patients with gastric cancer peritoneal carcinomatosis. World J Gastrointest Oncol. 2018; 10:282–289.
Article2. Thomassen I, van Gestel YR, van Ramshorst B, Luyer MD, Bosscha K, Nienhuijs SW, et al. Peritoneal carcinomatosis of gastric origin: a population-based study on incidence, survival and risk factors. Int J Cancer. 2014; 134:622–628.
Article3. Maeda H, Kobayashi M, Sakamoto J. Evaluation and treatment of malignant ascites secondary to gastric cancer. World J Gastroenterol. 2015; 21:10936–10947.
Article4. Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009; 136:215–233.
Article5. Lin S, Gregory RI. MicroRNA biogenesis pathways in cancer. Nat Rev Cancer. 2015; 15:321–333.
Article6. Cortez MA, Bueso-Ramos C, Ferdin J, Lopez-Berestein G, Sood AK, Calin GA. MicroRNAs in body fluids--the mix of hormones and biomarkers. Nat Rev Clin Oncol. 2011; 8:467–477.
Article7. Ahadi M, Tehranian S, Memar B, Vossoughinia H, Salari M, Eskandari E, et al. Diagnostic value of carcinoembryonic antigen in malignancy-related ascites: systematic review and meta-analysis. Acta Gastroenterol Belg. 2014; 77:418–424.8. Han ES, Lee HH, Lee JS, Song KY, Park CH, Jeon HM. At which stage of gastric cancer progression do levels of carcinoembryonic antigen and carbohydrate antigen 19-9 increase? Application in advanced gastric cancer treatment. J Gastric Cancer. 2014; 14:123–128.
Article9. Helwa I, Cai J, Drewry MD, Zimmerman A, Dinkins MB, Khaled ML, et al. A comparative study of serum exosome isolation using differential ultracentrifugation and three commercial reagents. PLoS One. 2017; 12:e0170628.
Article10. Markowska A, Pendergrast RS, Pendergrast JS, Pendergrast PS. A novel method for the isolation of extracellular vesicles and RNA from urine. J Circ Biomark. 2017; 6:1849454417712666.
Article11. Jamel S, Markar SR, Malietzis G, Acharya A, Athanasiou T, Hanna GB. Prognostic significance of peritoneal lavage cytology in staging gastric cancer: systematic review and meta-analysis. Gastric Cancer. 2018; 21:10–18.
Article12. Guideline Committee of the Korean Gastric Cancer Association (KGCA), Development Working Group & Review Panel. Korean practice guideline for gastric cancer 2018: an evidence-based, multi-disciplinary approach. J Gastric Cancer. 2019; 19:1–48.13. Vaksman O, Stavnes HT, Kaern J, Trope CG, Davidson B, Reich R. miRNA profiling along tumour progression in ovarian carcinoma. J Cell Mol Med. 2011; 15:1593–1602.
Article14. Vaksman O, Tropé C, Davidson B, Reich R. Exosome-derived miRNAs and ovarian carcinoma progression. Carcinogenesis. 2014; 35:2113–2120.
Article15. Tokuhisa M, Ichikawa Y, Kosaka N, Ochiya T, Yashiro M, Hirakawa K, et al. Exosomal miRNAs from peritoneum lavage fluid as potential prognostic biomarkers of peritoneal metastasis in gastric cancer. PLoS One. 2015; 10:e0130472.
Article16. Yamamoto CM, Oakes ML, Murakami T, Muto MG, Berkowitz RS, Ng SW. Comparison of benign peritoneal fluid- and ovarian cancer ascites-derived extracellular vesicle RNA biomarkers. J Ovarian Res. 2018; 11:20.
Article17. Záveský L, Jandáková E, Weinberger V, Minář L, Hanzíková V, Dušková D, et al. Ascites-derived extracellular microRNAs as potential biomarkers for ovarian cancer. Reprod Sci. 2019; 26:510–522.
Article18. Schindler P, Kupcinskas J, Juzenas S, Skieceviciene J, Salteniene V, Schulz C, et al. Expression of microRNAs in the ascites of patients with peritoneal carcinomatosis and peritonitis. Cancer Cytopathol. 2018; 126:353–363.
Article19. Kaleta EJ, Tolan NV, Ness KA, O'Kane D, Algeciras-Schimnich A. CEA, AFP and CA 19-9 analysis in peritoneal fluid to differentiate causes of ascites formation. Clin Biochem. 2013; 46:814–818.
Article20. Zhu FL, Ling AS, Wei Q, Ma J, Lu G. Tumor markers in serum and ascites in the diagnosis of benign and malignant ascites. Asian Pac J Cancer Prev. 2015; 16:719–722.
Article21. Chubb SP, Williams RA. Biochemical analysis of pleural fluid and ascites. Clin Biochem Rev. 2018; 39:39–50.22. Jung M, Jeung HC, Lee SS, Park JY, Hong S, Lee SH, et al. The clinical significance of ascitic fluid CEA in advanced gastric cancer with ascites. J Cancer Res Clin Oncol. 2010; 136:517–526.
Article23. Song SE, Choi P, Kim JH, Jung K, Kim SE, Moon W, et al. Diagnostic value of carcinoembryonic antigen in ascites for colorectal cancer with peritoneal carcinomatosis. Korean J Gastroenterol. 2018; 71:332–337.
Article24. Tian F, Shen Y, Chen Z, Li R, Lu J, Ge Q. Aberrant miR-181b-5p and miR-486-5p expression in serum and tissue of non-small cell lung cancer. Gene. 2016; 591:338–343.
Article25. Nakajima G, Hayashi K, Xi Y, Kudo K, Uchida K, Takasaki K, et al. Non-coding microRNAs hsa-let-7g and hsa-miR-181b are associated with chemoresponse to S-1 in colon cancer. Cancer Genomics Proteomics. 2006; 3:317–324.26. Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, et al. MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One. 2014; 9:e96228.
Article27. Shi ZM, Wang XF, Qian X, Tao T, Wang L, Chen QD, et al. miRNA-181b suppresses IGF-1R and functions as a tumor suppressor gene in gliomas. RNA. 2013; 19:552–560.
Article28. Ueda T, Volinia S, Okumura H, Shimizu M, Taccioli C, Rossi S, et al. Relation between microRNA expression and progression and prognosis of gastric cancer: a microRNA expression analysis. Lancet Oncol. 2010; 11:136–146.
Article29. Jiang J, Zheng X, Xu X, Zhou Q, Yan H, Zhang X, et al. Prognostic significance of miR-181b and miR-21 in gastric cancer patients treated with S-1/Oxaliplatin or Doxifluridine/Oxaliplatin. PLoS One. 2011; 6:e23271.
Article30. Saito M, Okayama H, Saito K, Ando J, Kumamoto K, Nakamura I, et al. CDX2 is involved in microRNA-associated inflammatory carcinogenesis in gastric cancer. Oncol Lett. 2017; 14:6184–6190.
Article31. Zhou Q, Zheng X, Chen L, Xu B, Yang X, Jiang J, et al. Smad2/3/4 pathway contributes to TGF-β-induced miRNA-181b expression to promote gastric cancer metastasis by targeting Timp3. Cell Physiol Biochem. 2016; 39:453–466.
Article32. Li X, Zhang Y, Zhang H, Liu X, Gong T, Li M, et al. miRNA-223 promotes gastric cancer invasion and metastasis by targeting tumor suppressor EPB41L3. Mol Cancer Res. 2011; 9:824–833.
Article33. Huang S, Wang J, Li J, Luo Q, Zhao M, Zheng L, et al. Serum microRNA expression profile as a diagnostic panel for gastric cancer. Jpn J Clin Oncol. 2016; 46:811–818.
Article34. Chen L, Yang Q, Kong WQ, Liu T, Liu M, Li X, et al. MicroRNA-181b targets cAMP responsive element binding protein 1 in gastric adenocarcinomas. IUBMB Life. 2012; 64:628–635.
Article35. Li LQ, Yang Y, Chen H, Zhang L, Pan D, Xie WJ. MicroRNA-181b inhibits glycolysis in gastric cancer cells via targeting hexokinase 2 gene. Cancer Biomark. 2016; 17:75–81.
Article36. Wang B, Li W, Guo K, Xiao Y, Wang Y, Fan J. miR-181b promotes hepatic stellate cells proliferation by targeting p27 and is elevated in the serum of cirrhosis patients. Biochem Biophys Res Commun. 2012; 421:4–8.
Article37. Vuppalanchi R, Liang T, Goswami CP, Nalamasu R, Li L, Jones D, et al. Relationship between differential hepatic microRNA expression and decreased hepatic cytochrome P450 3A activity in cirrhosis. PLoS One. 2013; 8:e74471.
Article38. Gui J, Tian Y, Wen X, Zhang W, Zhang P, Gao J, et al. Serum microRNA characterization identifies miR-885-5p as a potential marker for detecting liver pathologies. Clin Sci (Lond). 2011; 120:183–193.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Exosomal CircPRRX1 Enhances Doxorubicin Resistance in Gastric Cancer by Regulating MiR-3064-5p/PTPN14 Signaling
- Combined Detection of Serum MiR-221-3p and MiR-122-5p Expression in Diagnosis and Prognosis of Gastric Cancer
- Urine exosomal bkv-miR-B1-5p and BK virus nephropathy in kidney transplant recipients
- Expression of specific microRNAs in tissue and plasma in colorectal cancer
- Malignant Ascites after Subduroperitoneal Shunt in a Patient with Leptomeningeal Metastasis