J Breast Cancer.
2013 Mar;16(1):23-31.
Expression of DNA Methyltransferases in Breast Cancer Patients and to Analyze the Effect of Natural Compounds on DNA Methyltransferases and Associated Proteins
- Affiliations
-
- 1Department of Biochemistry, All India Institute of Medical Sciences, New Delhi, India. ralhanr@gmail.com
- 2Dev Sanskriti Vishwa Vidhyalaya, Hardwar, India.
- 3Department of Surgery, All India Institute of Medical Sciences, New Delhi, India.
- 4Department of Pathology, All India Institute of Medical Sciences, New Delhi, India.
- 5Alex and Simona Shnaider Laboratory of Molecular Oncology, Mount Sinai Hospital, Toronto, Canada.
- 6Department of Pathology and Laboratory Medicine, Mount Sinai Hospital, Toronto, Canada.
- 7Sonshine Family Centre for Head & Neck Diseases, Mount Sinai Hospital, Toronto, Canada.
- 8Department of Otolaryngology, Mount Sinai Hospital, Toronto, Canada.
Abstract
- PURPOSE
The DNA methylation mediated by specific DNA methyltransferases (DNMTs), results in the epigenetic silencing of multiple genes which are implicated in human breast cancer. We hypothesized that the natural compounds modulate the expression of DNMTs and their associated proteins in the breast cancer cell lines and affect the methylation mediated gene silencing.
METHODS
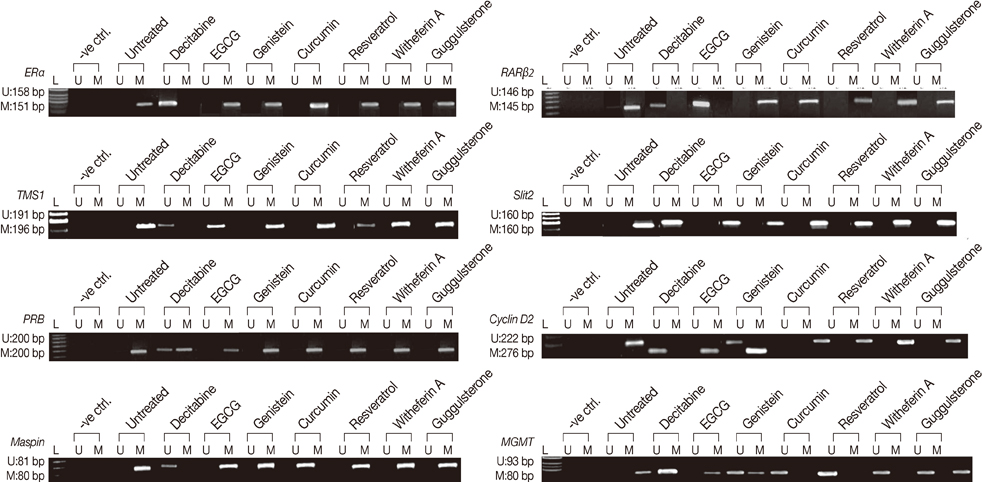
The DNMTs transcript expression was analyzed by reverse transcription-polymerase chain reaction (RT-PCR) in the tumors and the adjacent normal breast tissues of the patients with invasive ductal breast carcinoma. We tested the hypothesis that the natural compounds, viz., epigallocatechin gallate (EGCG), genistein, withaferin A, curcumin, resveratrol, and guggulsterone, have demethylation potential. To investigate this hypothesis, we analyzed the DNMTs expression at the transcript levels, followed by the analysis of DNMT1 and its associated proteins (HDAC1, MeCP2, and MBD2).
RESULTS
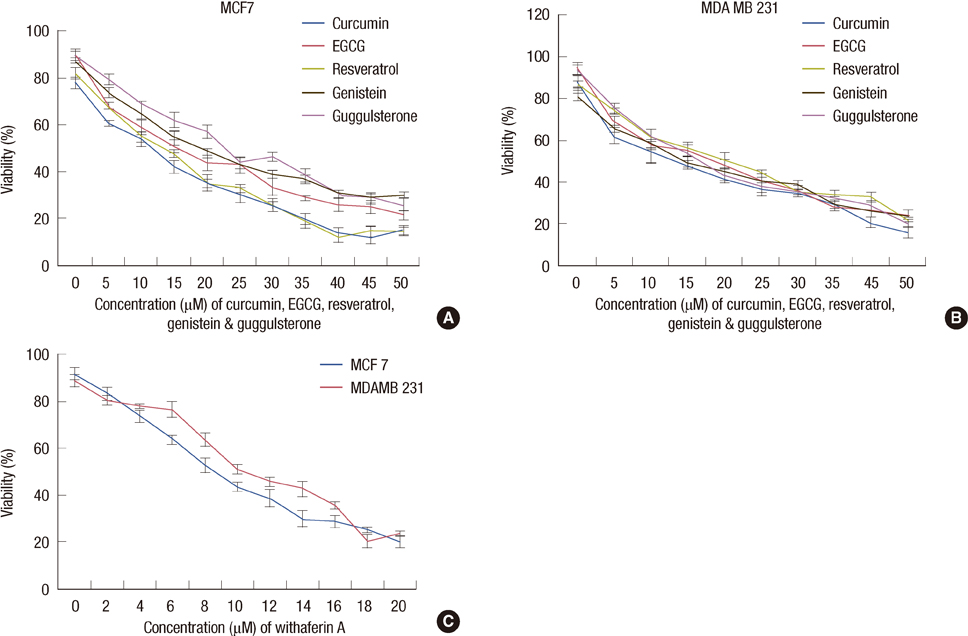
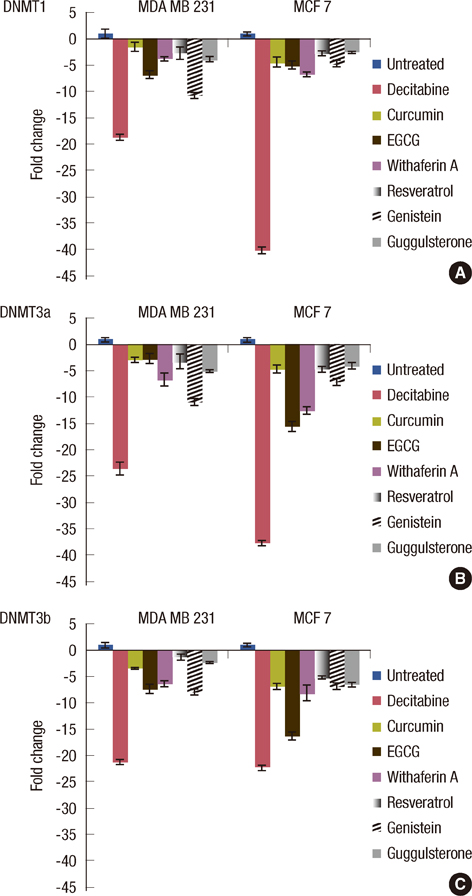
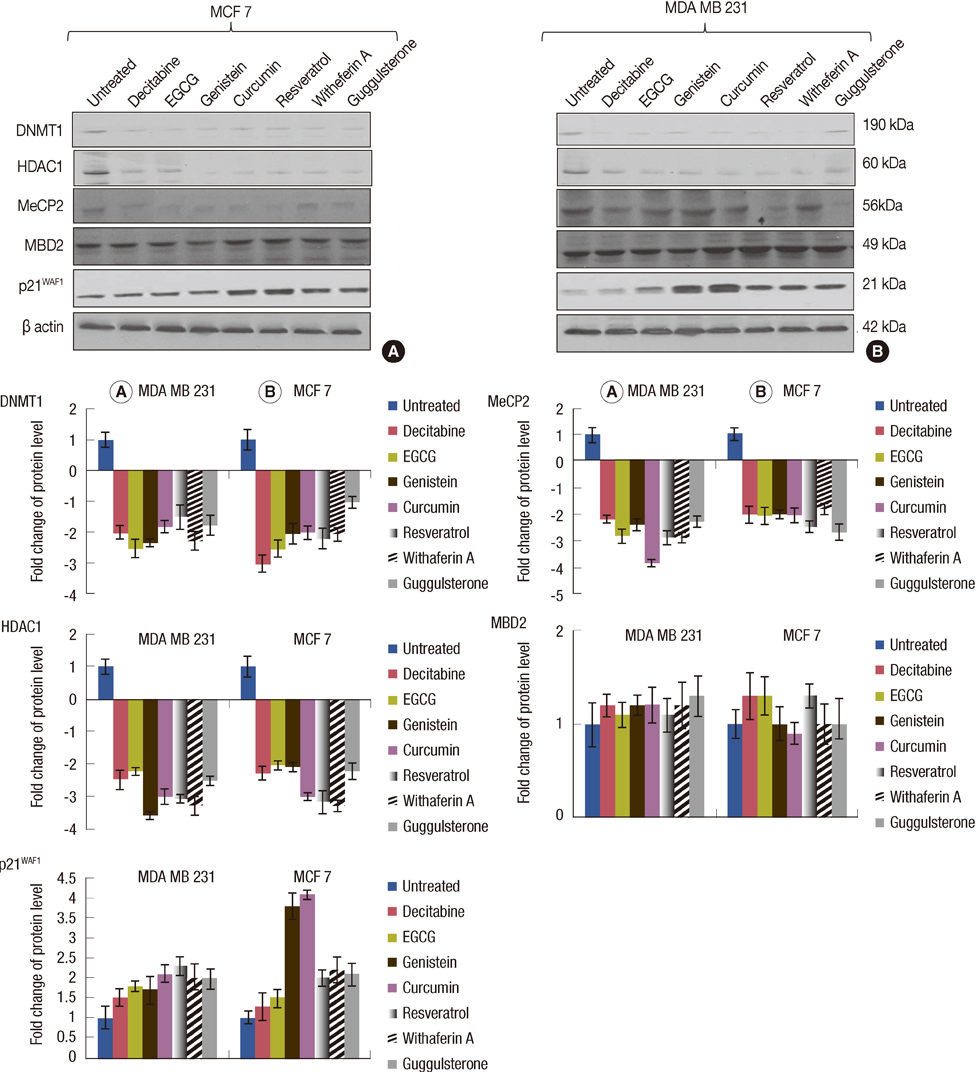
The increased DNMTs transcripts expression, viz., DNMT1, DNMT3a, and DNMT3b, in the breast cancer tissues suggest involvement of the DNMTs in the breast carcinogenesis. Quantitative RT-PCR analysis revealed that the treatment with natural compounds, viz., EGCG, genistein, withaferin A, curcumin, resveratrol, and guggulsterone, resulted in a significant decrease in the transcript levels of all the DNMTs investigated. Importantly, these natural compounds decreased the protein levels of DNMT1, HDAC1, and MeCP2.
CONCLUSION
Our results demonstrate that the natural compounds, EGCG, genistein, withaferin A, curcumin, resveratrol, and guggulsterone, have the potential to reverse the epigenetic changes. Moreover, their lack of toxicity makes these natural compounds promising candidates for the chemoprevention of the breast cancer. In-depth future mechanistic studies aimed to elucidate how these compounds affect the gene transcription are warranted.
MeSH Terms
Figure
Reference
-
1. Jones PA. DNA methylation and cancer. Oncogene. 2002. 21:5358–5360.
Article2. Ptak C, Petronis A. Epigenetics and complex disease: from etiology to new therapeutics. Annu Rev Pharmacol Toxicol. 2008. 48:257–276.
Article3. Siedlecki P, Zielenkiewicz P. Mammalian DNA methyltransferases. Acta Biochim Pol. 2006. 53:245–256.
Article4. Lyko F, Brown R. DNA methyltransferase inhibitors and the development of epigenetic cancer therapies. J Natl Cancer Inst. 2005. 97:1498–1506.
Article5. Fang MZ, Wang Y, Ai N, Hou Z, Sun Y, Lu H, et al. Tea polyphenol (-)-epigallocatechin-3-gallate inhibits DNA methyltransferase and reactivates methylation-silenced genes in cancer cell lines. Cancer Res. 2003. 63:7563–7570.6. Fini L, Selgrad M, Fogliano V, Graziani G, Romano M, Hotchkiss E, et al. Annurca apple polyphenols have potent demethylating activity and can reactivate silenced tumor suppressor genes in colorectal cancer cells. J Nutr. 2007. 137:2622–2628.
Article7. King-Batoon A, Leszczynska JM, Klein CB. Modulation of gene methylation by genistein or lycopene in breast cancer cells. Environ Mol Mutagen. 2008. 49:36–45.
Article8. Liu Z, Xie Z, Jones W, Pavlovicz RE, Liu S, Yu J, et al. Curcumin is a potent DNA hypomethylation agent. Bioorg Med Chem Lett. 2009. 19:706–709.
Article9. Majid S, Dar AA, Ahmad AE, Hirata H, Kawakami K, Shahryari V, et al. BTG3 tumor suppressor gene promoter demethylation, histone modification and cell cycle arrest by genistein in renal cancer. Carcinogenesis. 2009. 30:662–670.
Article10. Ramos S. Effects of dietary flavonoids on apoptotic pathways related to cancer chemoprevention. J Nutr Biochem. 2007. 18:427–442.
Article11. Choi JS, Choi YJ, Park SH, Kang JS, Kang YH. Flavones mitigate tumor necrosis factor-alpha-induced adhesion molecule upregulation in cultured human endothelial cells: role of nuclear factor-kappa B. J Nutr. 2004. 134:1013–1019.
Article12. Sim GS, Lee BC, Cho HS, Lee JW, Kim JH, Lee DH, et al. Structure activity relationship of antioxidative property of flavonoids and inhibitory effect on matrix metalloproteinase activity in UVA-irradiated human dermal fibroblast. Arch Pharm Res. 2007. 30:290–298.
Article13. Lee WJ, Shim JY, Zhu BT. Mechanisms for the inhibition of DNA methyltransferases by tea catechins and bioflavonoids. Mol Pharmacol. 2005. 68:1018–1030.
Article14. Lee WJ, Zhu BT. Inhibition of DNA methylation by caffeic acid and chlorogenic acid, two common catechol-containing coffee polyphenols. Carcinogenesis. 2006. 27:269–277.
Article15. Chen D, Milacic V, Chen MS, Wan SB, Lam WH, Huo C, et al. Tea polyphenols, their biological effects and potential molecular targets. Histol Histopathol. 2008. 23:487–496.16. Chen D, Wang CY, Lambert JD, Ai N, Welsh WJ, Yang CS. Inhibition of human liver catechol-O-methyltransferase by tea catechins and their metabolites: structure-activity relationship and molecular-modeling studies. Biochem Pharmacol. 2005. 69:1523–1531.
Article17. Moiseeva EP, Almeida GM, Jones GD, Manson MM. Extended treatment with physiologic concentrations of dietary phytochemicals results in altered gene expression, reduced growth, and apoptosis of cancer cells. Mol Cancer Ther. 2007. 6:3071–3079.
Article18. Mirza S, Sharma G, Prasad CP, Parshad R, Srivastava A, Gupta SD, et al. Promoter hypermethylation of TMS1, BRCA1, ERalpha and PRB in serum and tumor DNA of invasive ductal breast carcinoma patients. Life Sci. 2007. 81:280–287.
Article19. Sharma G, Mirza S, Prasad CP, Srivastava A, Gupta SD, Ralhan R. Promoter hypermethylation of p16INK4A, p14ARF, CyclinD2 and Slit2 in serum and tumor DNA from breast cancer patients. Life Sci. 2007. 80:1873–1881.
Article20. Shukla S, Mirza S, Sharma G, Parshad R, Gupta SD, Ralhan R. Detection of RASSF1A and RARbeta hypermethylation in serum DNA from breast cancer patients. Epigenetics. 2006. 1:88–93.
Article21. Garinis GA, Patrinos GP, Spanakis NE, Menounos PG. DNA hypermethylation: when tumour suppressor genes go silent. Hum Genet. 2002. 111:115–127.
Article22. Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000. 16:168–174.
Article23. Esteller M. Epigenetics in cancer. N Engl J Med. 2008. 358:1148–1159.
Article24. Li Y, Tollefsbol TO. Impact on DNA methylation in cancer prevention and therapy by bioactive dietary components. Curr Med Chem. 2010. 17:2141–2151.
Article25. Rice JC, Allis CD. Histone methylation versus histone acetylation: new insights into epigenetic regulation. Curr Opin Cell Biol. 2001. 13:263–273.
Article26. Prokhortchouk E, Hendrich B. Methyl-CpG binding proteins and cancer: are MeCpGs more important than MBDs? Oncogene. 2002. 21:5394–5399.
Article27. Fang JY, Lu YY. Effects of histone acetylation and DNA methylation on p21( WAF1) regulation. World J Gastroenterol. 2002. 8:400–405.
Article28. Periyasamy S, Ammanamanchi S, Tillekeratne MP, Brattain MG. Repression of transforming growth factor-beta receptor type I promoter expression by Sp1 deficiency. Oncogene. 2000. 19:4660–4667.
Article29. Berger J, Bird A. Role of MBD2 in gene regulation and tumorigenesis. Biochem Soc Trans. 2005. 33(Pt 6):1537–1540.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Effects of β-carotene on Expression of Selected MicroRNAs, Histone Acetylation, and DNA Methylation in Colon Cancer Stem Cells
- Epigenetic Changes (Aberrant DNA Methylation) in Colorectal Neoplasia
- DNA ploidy, S-phase activity and c-erbB-2 oncogene protein expression in breast cancer and its relationship to prognosis
- Inhibitors of DNA methylation support TGF-β1-induced IL11 expression in gingival fibroblasts
- Embryonic Stem Cells Lacking DNA Methyltransferases Differentiate into Neural Stem Cells that Are Defective in Self-Renewal