J Vet Sci.
2016 Mar;17(1):71-78. 10.4142/jvs.2016.17.1.71.
A novel M2e-multiple antigenic peptide providing heterologous protection in mice
- Affiliations
-
- 1Division of Swine Infectious Diseases, Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai 200241, China. gztong@shvri.ac.cn
- KMID: 2363337
- DOI: http://doi.org/10.4142/jvs.2016.17.1.71
Abstract
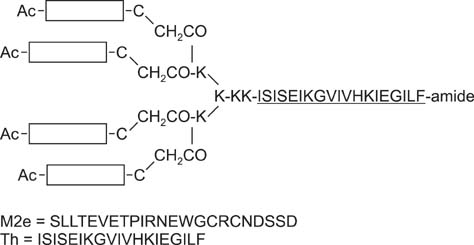
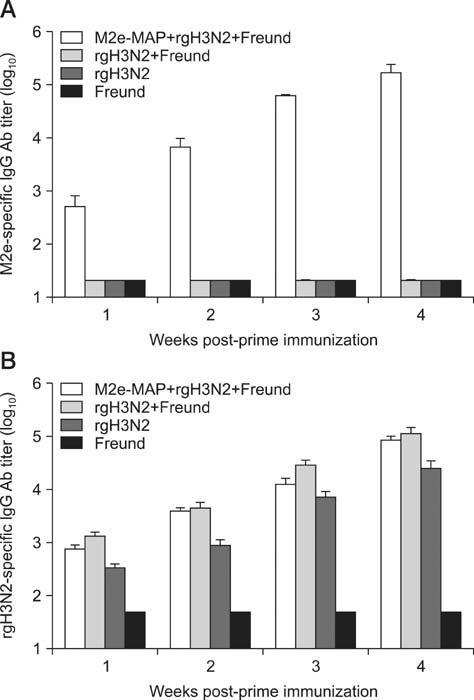
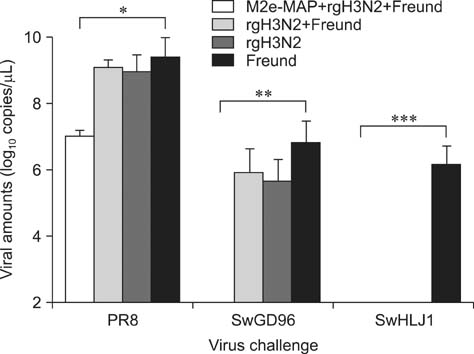
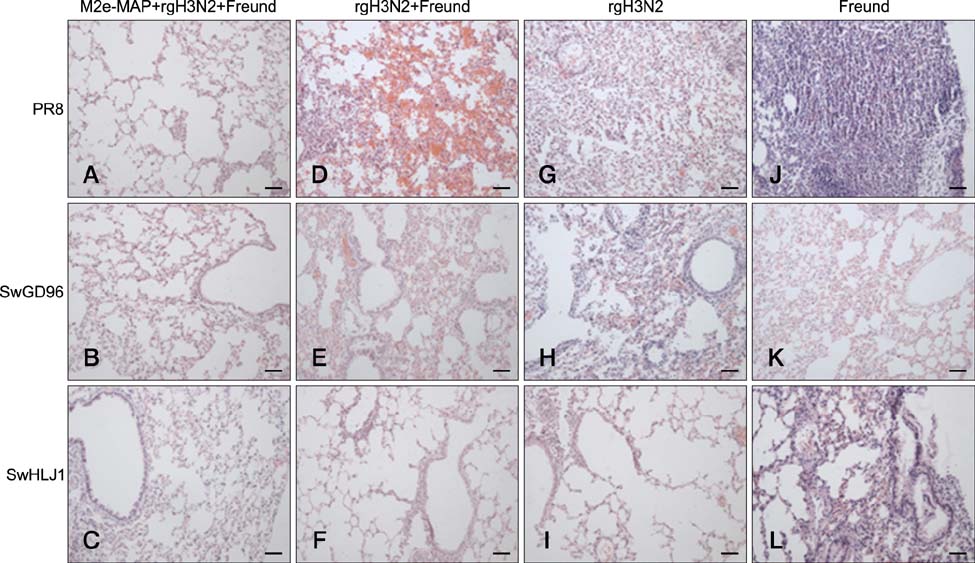
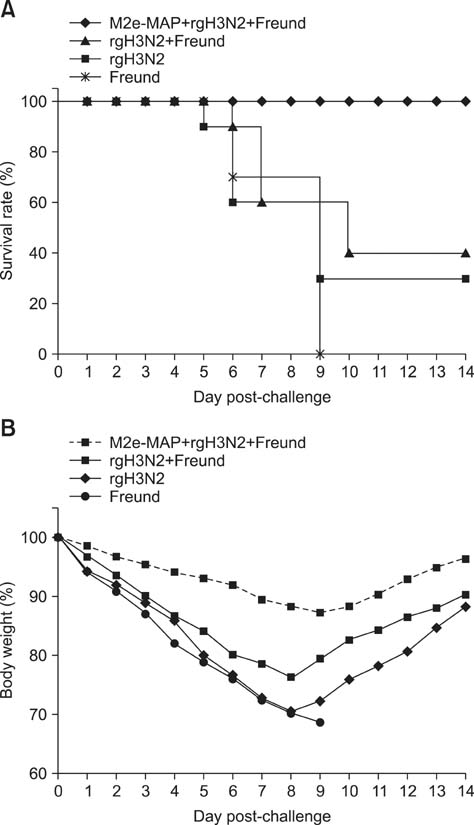
- Swine influenza viruses (SwIVs) cause considerable morbidity and mortality in domestic pigs, resulting in a significant economic burden. Moreover, pigs have been considered to be a possible mixing vessel in which novel strains loom. Here, we developed and evaluated a novel M2e-multiple antigenic peptide (M2e-MAP) as a supplemental antigen for inactivated H3N2 vaccine to provide cross-protection against two main subtypes of SwIVs, H1N1 and H3N2. The novel tetra-branched MAP was constructed by fusing four copies of M2e to one copy of foreign T helper cell epitopes. A high-yield reassortant H3N2 virus was generated by plasmid based reverse genetics. The efficacy of the novel H3N2 inactivated vaccines with or without M2e-MAP supplementation was evaluated in a mouse model. M2e-MAP conjugated vaccine induced strong antibody responses in mice. Complete protection against the heterologous swine H1N1 virus was observed in mice vaccinated with M2e-MAP combined vaccine. Moreover, this novel peptide confers protection against lethal challenge of A/Puerto Rico/8/34 (H1N1). Taken together, our results suggest the combined immunization of reassortant inactivated H3N2 vaccine and the novel M2e-MAP provided cross-protection against swine and human viruses and may serve as a promising approach for influenza vaccine development.
Keyword
MeSH Terms
-
Animals
Antibodies, Viral/blood
Antigens, Viral/genetics/*immunology
Body Weight
Cross Protection/*immunology
Disease Models, Animal
Epitopes, T-Lymphocyte/genetics/immunology
Female
Influenza A Virus, H3N2 Subtype/genetics/*immunology
Influenza Vaccines/*immunology
Mice
Mice, Inbred BALB C
Orthomyxoviridae Infections/*immunology/mortality/pathology/prevention & control
Peptides/genetics/*immunology
Random Allocation
Survival Analysis
Vaccines, Synthetic/immunology
Virus Replication
Antibodies, Viral
Antigens, Viral
Epitopes, T-Lymphocyte
Influenza Vaccines
Peptides
Vaccines, Synthetic
Figure
Reference
-
1. Carragher DM, Kaminski DA, Moquin A, Hartson L, Randall TD. A novel role for non-neutralizing antibodies against nucleoprotein in facilitating resistance to influenza virus. J Immunol. 2008; 181:4168–4176.
Article2. Center for Disease Control and Prevention (US). Update: Influenza A (H3N2)v transmission and guidelines - five states, 2011. MMWR Morb Mortal Wkly Rep. 2012; 60:1741–1744.3. El Bakkouri K, Descamps F, De Filette M, Smet A, Festjens E, Birkett A, Van Rooijen N, Verbeek S, Fiers W, Saelens X. Universal vaccine based on ectodomain of matrix protein 2 of influenza A: Fc receptors and alveolar macrophages mediate protection. J Immunol. 2011; 186:1022–1031.
Article4. Fiers W, De Filette M, Birkett A, Neirynck S, Min Jou W. A "universal" human influenza A vaccine. Virus Res. 2004; 103:173–176.
Article5. Gillim-Ross L, Subbarao K. Emerging respiratory viruses: challenges and vaccine strategies. Clin Microbiol Rev. 2006; 19:614–636.
Article6. Gioia C, Agrati C, Castilletti C, Capobianchi MR, Martini F. Influenza pandemics, immune cross-reactivity, and pandemic control strategies. J Infect Dis. 2008; 198:294–295. author reply 295-296.
Article7. Gioia C, Castilletti C, Tempestilli M, Piacentini P, Bordi L, Chiappini R, Agrati C, Squarcione S, Ippolito G, Puro V, Capobianchi MR, Poccia F. Cross-subtype immunity against avian influenza in persons recently vaccinated for influenza. Emerg Infec Dis. 2008; 14:121–128.
Article8. Hughey PG, Roberts PC, Holsinger LJ, Zebedee SL, Lamb RA, Compans RW. Effects of antibody to the influenza A virus M2 protein on M2 surface expression and virus assembly. Virology. 1995; 212:411–421.
Article9. Jegerlehner A, Schmitz N, Storni T, Bachmann MF. Influenza A vaccine based on the extracellular domain of M2: weak protection mediated via antibody-dependent NK cell activity. J Immunol. 2004; 172:5598–5605.
Article10. Kida H, Brown LE, Webster RG. Biological activity of monoclonal antibodies to operationally defined antigenic regions on the hemagglutinin molecule of A/Seal/Massachusetts/1/80 (H7N7) influenza virus. Virology. 1982; 122:38–47.
Article11. Lu X, Tumpey TM, Morken T, Zaki SR, Cox NJ, Katz JM. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol. 1999; 73:5903–5911.
Article12. Ma JH, Yang FR, Yu H, Zhou YJ, Li GX, Huang M, Wen F, Tong G. An M2e-based synthetic peptide vaccine for influenza A virus confers heterosubtypic protection from lethal virus challenge. Virol J. 2013; 10:227.
Article13. Nguyen HH, Moldoveanu Z, Novak MJ, van Ginkel FW, Ban E, Kiyono H, McGhee JR, Mestecky J. Heterosubtypic immunity to lethal influenza A virus infection is associated with virus-specific CD8+ cytotoxic T lymphocyte responses induced in mucosa-associated tissues. Virology. 1999; 254:50–60.
Article14. Olsen CW, Karasin AI, Carman S, Li Y, Bastien N, Ojkic D, Alves D, Charbonneau G, Henning BM, Low DE, Burton L, Broukhanski G. Triple reassortant H3N2 influenza A viruses, Canada, 2005. Emerg Infect Dis. 2006; 12:1132–1135.
Article15. Park JK, Lee DH, Cho CH, Yuk SS, To EO, Kwon JH, Noh JY, Kim BY, Choi SW, Shim BS, Song MK, Lee JB, Park SY, Choi IS, Song CS. Supplementation of oil-based inactivated H9N2 vaccine with M2e antigen enhances resistance against heterologous H9N2 avian influenza virus infection. Vet Microbiol. 2014; 169:211–217.
Article16. Roberts PC, Lamb RA, Compans RW. The M1 and M2 proteins of influenza A virus are important determinants in filamentous particle formation. Virology. 1998; 240:127–137.
Article17. Shortridge KF, Webster RG, Butterfield WK, Campbell CH. Persistence of Hong Kong influenza virus variants in pigs. Science. 1977; 196:1454–1455.
Article18. Tompkins SM, Zhao ZS, Lo CY, Misplon JA, Liu T, Ye Z, Hogan RJ, Wu Z, Benton KA, Tumpey TM, Epstein SL. Matrix protein 2 vaccination and protection against influenza viruses, including subtype H5N1. Emerg Infect Dis. 2007; 13:426–435.
Article19. Vincent AL, Ma W, Lager KM, Janke BH, Richt JA. Swine influenza viruses a North American perspective. Adv Virus Res. 2008; 72:127–154.20. Wen F, Ma JH, Yu H, Yang FR, Huang M, Zhou YJ, Li ZJ, Tong GZ. Protective efficacy of a high-growth reassortant swine H3N2 inactivated vaccine constructed by reverse genetic manipulation. J Vet Sci. 2014; 15:381–388.
Article21. Wen F, Yu H, Yang FR, Huang M, Yang S, Zhou YJ, Li ZJ, Tong GZ. Efficacy of a high-growth reassortant H1N1 influenza virus vaccine against the classical swine H1N1 subtype influenza virus in mice and pigs. Arch Virol. 2014; 159:2957–2967.
Article22. Yu H, Hua RH, Zhang Q, Liu TQ, Liu HL, Li GX, Tong GZ. Genetic evolution of swine influenza A (H3N2) viruses in China from 1970 to 2006. J Clin Microbiol. 2008; 46:1067–1075.
Article23. Zebedee SL, Lamb RA. Influenza A virus M2 protein: monoclonal antibody restriction of virus growth and detection of M2 in virions. J Virol. 1988; 62:2762–2772.
Article24. Zhao G, Lin Y, Du L, Guan J, Sun S, Sui H, Kou Z, Chan CC, Guo Y, Jiang S, Zheng BJ, Zhou Y. An M2e-based multiple antigenic peptide vaccine protects mice from lethal challenge with divergent H5N1 influenza viruses. Virol J. 2010; 7:9.
Article25. Zhou NN, Senne DA, Landgraf JS, Swenson SL, Erickson G, Rossow K, Liu L, Yoon K, Krauss S, Webster RG. Genetic reassortment of avian, swine, and human influenza A viruses in American pigs. J Virol. 1999; 73:8851–8856.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Evaluation of Protective Immunity of Peptide Vaccines Composed of a 15-mer N-terminal Matrix Protein 2 and a Helper T-Cell Epitope Derived from Influenza A Virus
- Mechanisms of Cross-protection by Influenza Virus M2-based Vaccines
- Imunological characterization of antigens from cysticercus and sparganum and their application to immunodiagnosis 1. Immunological characteristics of crude antigenic components from Cysticercus cellulosae
- T Cells Modified with CD70 as an Alternative Cellular Vaccine for Antitumor Immunity
- Longevity of Antibodies to Live Orientia tsutsugamushi Inoculated in Sprague Dawley Rats