Hanyang Med Rev.
2013 May;33(2):83-89. 10.7599/hmr.2013.33.2.83.
Autophagy in Redox Signalling
- Affiliations
-
- 1Department of Biochemistry and Molecular Biology, School of Medicine, Kyung Hee University, Seoul, Korea. sgskim@khu.ac.kr
- KMID: 2168235
- DOI: http://doi.org/10.7599/hmr.2013.33.2.83
Abstract
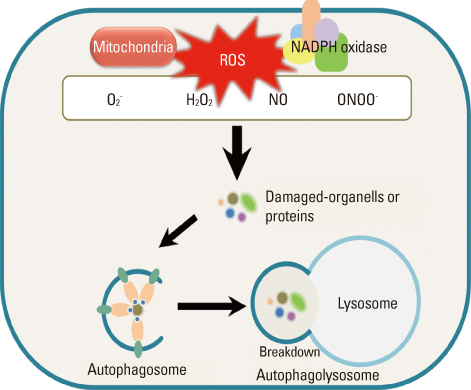
- Autophagy is a major catabolic process that is involved in cellular degradation of unnecessary or dysfunctional cellular components via the lysosomal machinery. Autophagy is involved in a variety of biological processes such as programmed cell death, removal of damaged organelles and development of different tissue-specific functions. In recent experiments, the role of autophagy as an important mediator of the pathological response to redox signalling of cellular damage has been elucidated and expanded. Oxidative stresses to the cellular system induces autophagy as a means to selectively remove oxidatively modified macromolecules and organelles. Reactive oxygen species (ROS) are highly reactive oxygen free radicals that are produced as by-products of cellular metabolism, primarily by mitochondria and NADPH oxidases. ROS can be beneficial or harmful to cells and tissues by depending on their concentration and location. ROS function as redox messengers in intracellular signalling at physiologically low level, whereas excess ROS can induce oxidative modification of cellular macromolecules and eventually promote cell death. Thus, the interface of autophagy-related oxidative stress adaptation and cell death is important for understanding redox biology and pathogenesis. In this review, we describe the basic mechanism and function of autophagy in the context of response to oxidative stress and redox signalling in pathogenesis.
MeSH Terms
Figure
Cited by 1 articles
-
Do Reactive Oxygen Species Cause Aging?
Seong Eon Ryu
Hanyang Med Rev. 2013;33(2):75-76. doi: 10.7599/hmr.2013.33.2.75.
Reference
-
1. Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004; 14:70–77.
Article2. Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev Cell. 2004; 6:463–477.3. Sridhar S, Botbol Y, Macian F, Cuervo AM. Autophagy and disease: always two sides to a problem. J Pathol. 2012; 226:255–273.
Article4. Shacka JJ, Roth KA, Zhang J. The autophagy-lysosomal degradation pathway: role in neurodegenerative disease and therapy. Front Biosci. 2008; 13:718–736.
Article5. Gottlieb RA, Carreira RS. Autophagy in health and disease. 5. Mitophagy as a way of life. Am J Physiol Cell Physiol. 2010; 299:C203–C210.
Article6. He C, Klionsky DJ. Regulation mechanisms and signaling pathways of autophagy. Annu Rev Genet. 2009; 43:67–93.
Article7. Chen Y, Klionsky DJ. The regulation of autophagy - unanswered questions. J Cell Sci. 2011; 124:161–170.
Article8. Kabeya Y, Mizushima N, Ueno T, Yamamoto A, Kirisako T, Noda T, et al. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 2000; 19:5720–5728.
Article9. Bjorkoy G, Lamark T, Brech A, Outzen H, Perander M, Overvatn A, et al. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J Cell Biol. 2005; 171:603–614.
Article10. Gueraud F, Atalay M, Bresgen N, Cipak A, Eckl PM, Huc L, et al. Chemistry and biochemistry of lipid peroxidation products. Free Radic Res. 2010; 44:1098–1124.
Article11. Murphy MP. How mitochondria produce reactive oxygen species. Biochem J. 2009; 417:1–13.
Article12. Scherz-Shouval R, Shvets E, Fass E, Shorer H, Gil L, Elazar Z. Reactive oxygen species are essential for autophagy and specifically regulate the activity of Atg4. EMBO J. 2007; 26:1749–1760.
Article13. Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003; 115:577–590.
Article14. Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011; 13:132–141.
Article15. Egan DF, Shackelford DB, Mihaylova MM, Gelino S, Kohnz RA, Mair W, et al. Phosphorylation of ULK1 (hATG1) by AMP-activated protein kinase connects energy sensing to mitophagy. Science. 2011; 331:456–461.
Article16. Emerling BM, Weinberg F, Snyder C, Burgess Z, Mutlu GM, Viollet B, et al. Hypoxic activation of AMPK is dependent on mitochondrial ROS but independent of an increase in AMP/ATP ratio. Free Radic Biol Med. 2009; 46:1386–1391.
Article17. Alexander A, Cai SL, Kim J, Nanez A, Sahin M, MacLean KH, et al. ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc Natl Acad Sci U S A. 2010; 107:4153–4158.
Article18. Sims GP, Rowe DC, Rietdijk ST, Herbst R, Coyle AJ. HMGB1 and RAGE in inflammation and cancer. Annu Rev Immunol. 2010; 28:367–388.
Article19. Tang D, Kang R, Cheh CW, Livesey KM, Liang X, Schapiro NE, et al. HMGB1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene. 2010; 29:5299–5310.
Article20. Bellot G, Garcia-Medina R, Gounon P, Chiche J, Roux D, Pouyssegur J, et al. Hypoxia-induced autophagy is mediated through hypoxia-inducible factor induction of BNIP3 and BNIP3L via their BH3 domains. Mol Cell Biol. 2009; 29:2570–2581.
Article21. Byun YJ, Kim SK, Kim YM, Chae GT, Jeong SW, Lee SB. Hydrogen peroxide induces autophagic cell death in C6 glioma cells via BNIP3-mediated suppression of the mTOR pathway. Neurosci Lett. 2009; 461:131–135.
Article22. Li Y, Wang Y, Kim E, Beemiller P, Wang CY, Swanson J, et al. Bnip3 mediates the hypoxia-induced inhibition on mammalian target of rapamycin by interacting with Rheb. J Biol Chem. 2007; 282:35803–35813.
Article23. Hanna RA, Quinsay MN, Orogo AM, Giang K, Rikka S, Gustafsson AB. Microtubule-associated protein 1 light chain 3 (LC3) interacts with Bnip3 protein to selectively remove endoplasmic reticulum and mitochondria via autophagy. J Biol Chem. 2012; 287:19094–19104.
Article24. Wasserman WW, Fahl WE. Functional antioxidant responsive elements. Proc Natl Acad Sci U S A. 1997; 94:5361–5366.25. Ishii T, Itoh K, Yamamoto M. Roles of Nrf2 in activation of antioxidant enzyme genes via antioxidant responsive elements. Methods Enzymol. 2002; 348:182–190.26. Itoh K, Wakabayashi N, Katoh Y, Ishii T, Igarashi K, Engel JD, et al. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999; 13:76–86.
Article27. Li PF, Dietz R, von Harsdorf R. p53 regulates mitochondrial membrane potential through reactive oxygen species and induces cytochrome c-independent apoptosis blocked by Bcl-2. EMBO J. 1999; 18:6027–6036.
Article28. Bensaad K, Tsuruta A, Selak MA, Vidal MN, Nakano K, Bartrons R, et al. TIGAR, a p53-inducible regulator of glycolysis and apoptosis. Cell. 2006; 126:107–120.
Article29. Bensaad K, Cheung EC, Vousden KH. Modulation of intracellular ROS levels by TIGAR controls autophagy. EMBO J. 2009; 28:3015–3026.
Article30. Crighton D, Wilkinson S, O'Prey J, Syed N, Smith P, Harrison PR, et al. DRAM, a p53-induced modulator of autophagy, is critical for apoptosis. Cell. 2006; 126:121–134.
Article31. Budanov AV. Stress-responsive sestrins link p53 with redox regulation and mammalian target of rapamycin signaling. Antioxid Redox Signal. 2011; 15:1679–1690.
Article32. McCray BA, Taylor JP. The role of autophagy in age-related neurodegeneration. Neurosignals. 2008; 16:75–84.
Article33. Gal J, Strom AL, Kwinter DM, Kilty R, Zhang J, Shi P, et al. Sequestosome 1/p62 links familial ALS mutant SOD1 to LC3 via an ubiquitin-independent mechanism. J Neurochem. 2009; 111:1062–1073.
Article34. Hara T, Nakamura K, Matsui M, Yamamoto A, Nakahara Y, Suzuki-Migishima R, et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006; 441:885–889.
Article35. Lee JH, Yu WH, Kumar A, Lee S, Mohan PS, Peterhoff CM, et al. Lysosomal proteolysis and autophagy require presenilin 1 and are disrupted by Alzheimer-related PS1 mutations. Cell. 2010; 141:1146–1158.
Article36. Jin S. Autophagy, mitochondrial quality control, and oncogenesis. Autophagy. 2006; 2:80–84.
Article37. Jung HS, Chung KW, Won Kim J, Kim J, Komatsu M, Tanaka K, et al. Loss of autophagy diminishes pancreatic beta cell mass and function with resultant hyperglycemia. Cell Metab. 2008; 8:318–324.
Article38. Petersen KF, Dufour S, Befroy D, Garcia R, Shulman GI. Impaired mitochondrial activity in the insulin-resistant offspring of patients with type 2 diabetes. N Engl J Med. 2004; 350:664–671.
Article39. Jung HS, Lee MS. Role of autophagy in diabetes and mitochondria. Ann N Y Acad Sci. 2010; 1201:79–83.
Article40. Nishino I, Fu J, Tanji K, Yamada T, Shimojo S, Koori T, et al. Primary LAMP-2 deficiency causes X-linked vacuolar cardiomyopathy and myopathy (Danon disease). Nature. 2000; 406:906–910.
Article41. Zhu H, Tannous P, Johnstone JL, Kong Y, Shelton JM, Richardson JA, et al. Cardiac autophagy is a maladaptive response to hemodynamic stress. J Clin Invest. 2007; 117:1782–1793.
Article42. Nakai A, Yamaguchi O, Takeda T, Higuchi Y, Hikoso S, Taniike M, et al. The role of autophagy in cardiomyocytes in the basal state and in response to hemodynamic stress. Nat Med. 2007; 13:619–624.
Article43. Huang C, Andres AM, Ratliff EP, Hernandez G, Lee P, Gottlieb RA. Preconditioning involves selective mitophagy mediated by Parkin and p62/SQSTM1. PLoS One. 2011; 6:e20975.
Article44. Deretic V. Multiple regulatory and effector roles of autophagy in immunity. Curr Opin Immunol. 2009; 21:53–62.
Article45. Orvedahl A, Levine B. Eating the enemy within: autophagy in infectious diseases. Cell Death Differ. 2009; 16:57–69.
Article46. Lee HK, Lund JM, Ramanathan B, Mizushima N, Iwasaki A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science. 2007; 315:1398–1401.
Article47. Jounai N, Takeshita F, Kobiyama K, Sawano A, Miyawaki A, Xin KQ, et al. The Atg5 Atg12 conjugate associates with innate antiviral immune responses. Proc Natl Acad Sci U S A. 2007; 104:14050–14055.
Article48. Dengjel J, Schoor O, Fischer R, Reich M, Kraus M, Muller M, et al. Autophagy promotes MHC class II presentation of peptides from intracellular source proteins. Proc Natl Acad Sci U S A. 2005; 102:7922–7927.
Article49. Miller BC, Zhao Z, Stephenson LM, Cadwell K, Pua HH, Lee HK, et al. The autophagy gene ATG5 plays an essential role in B lymphocyte development. Autophagy. 2008; 4:309–314.
Article50. Nedjic J, Aichinger M, Emmerich J, Mizushima N, Klein L. Autophagy in thymic epithelium shapes the T-cell repertoire and is essential for tolerance. Nature. 2008; 455:396–400.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Unbalanced Redox With Autophagy in Cardiovascular Disease
- Neuronal Autophagy: Characteristic Features and Roles in Neuronal Pathophysiology
- Autophagy in the placenta
- Functional interplay between non-canonical inflammasomes and autophagy in inflammatory responses and diseases
- The role of autophagy in cell proliferation and differentiation during tooth development