Cancer Res Treat.
2004 Aug;36(4):246-254.
Identification of Genes Involved in Liver Cancer Cell Growth Using an Antisense Library of Phage Genomic DNA
- Affiliations
-
- 1WelGENE Inc., Daegu, Korea. jonggu@kmu.ac.kr
- 2Department of Medical Genetic Engineering, Keimyung University School of Medicine, Dongsan Medical Center, Daegu, Korea.
- 3Department of Microbiology, College of Natural Sciences, Kyungpook National University, Daegu, Korea.
Abstract
- PURPOSE
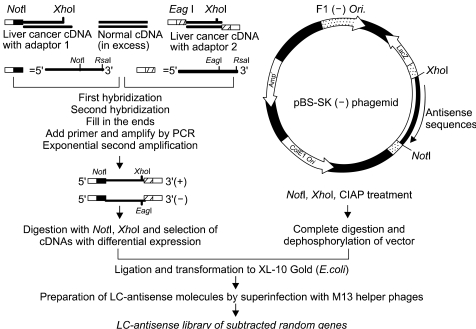
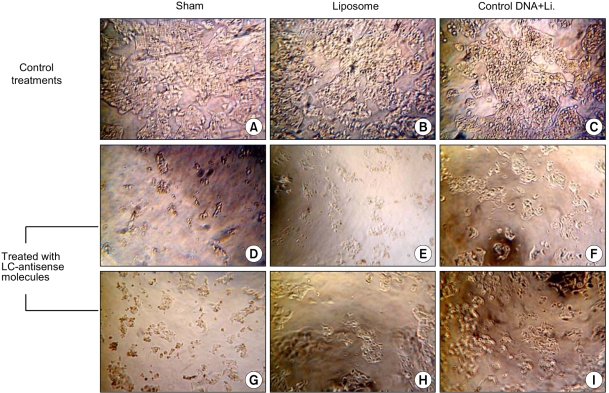
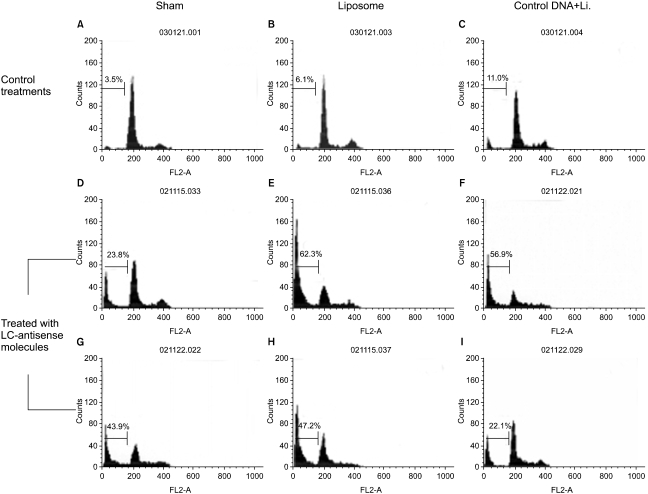
Genes involved in liver cancer cell growth have been identified using an antisense library of large circular (LC-) genomic DNA of a recombinant M13 phage. MATERIALS AND METHODS: A subtracted cDNA library was constructed by combining procedures of suppression subtractive hybridization (SSH) and unidirectional cloning of the subtracted cDNA into an M13 phagemid vector. Utilizing the life cycle of M13 bacteriophages, LC-antisense molecules derived from 1, 200 random cDNA clones selected by size were prepared from the culture supernatant of bacterial transformants. The antisense molecules were arrayed for transfection on 96-well plates preseeded with HepG2. RESULTS: When examined for growth inhibition after antisense transfection, 153 out of 1, 200 LC-antisense molecules showed varying degrees of growth inhibitory effect to HepG2 cells. Sequence comparison of the 153 clones identified 58 unique genes. The observations were further extended by other cell-based assays. CONCLUSION: These results suggest that the LC-antisense library offers potential for unique high-throughput screening to find genes involved in a specific biological function, and may prove to be an effective target validation system for gene-based drug discovery.
Keyword
MeSH Terms
Figure
Reference
-
1. Hieter P, Boguski M. Functional Genomics: It's all how you read it. Science. 1997; 278:601–602. PMID: 9381168.
Article3. Pandey A, Mann M. Proteomics to study genes and genomes. Nature. 2000; 405:837–846. PMID: 10866210.
Article4. Quackenbush J, Cho J, Lee D, Liang F, Holt I, Karamycheva S, Parvizi B, Pertea G, Sultana R, White J. The TIGR Gene Indices: analysis of gene transcript sequences in highly sampled eukaryotic species. Nucleic Acids Res. 2001; 29:159–164. PMID: 11125077.
Article5. Velculescu VE, Zhang L, Vogelstein B, Kinzler KW. Serial analysis of gene expression. Science. 1995; 270:484–487. PMID: 7570003.
Article6. Prashar Y, Weissman SM. READS: a method for display of 3'-end fragments of restriction enzyme-digested cDNAs for analysis of differential gene expression. Methods Enzymol. 1999; 303:258–272. PMID: 10349649.7. Thompson CB, Challoner PB, Neiman PE, Groudine M. Levels of c-myc oncogene mRNA are invariant throughout the cell cycle. Nature. 1985; 314:363–366. PMID: 3982504.
Article8. Melani C, Rivoltini L, Parmiani G, Calabretta B, Colombo MP. Inhibition of proliferation by c-myb antisense oligodeoxynucleotides in colon adenocarcinoma cell lines that express c-myb. Cancer Res. 1991; 51:2897–2901. PMID: 2032228.9. Anfossi G, Gewirtz AM, Calabretta B. An oligomer complementary to c-myb-encoded mRNA inhibits proliferation of human myeloid leukemia cell lines. Proc Natl Acad Sci USA. 1989; 86:3379–3383. PMID: 2541445.10. Graham MJ, Crooke ST, Monteith DK, Cooper SR, Lemonidis KM, Stecker KK, Martin MJ, Crooke RM. In vivo distribution and metabolism of a phosphorothioate oligonucleotide within rat liver after intravenous administration. J Pharmacol Exp Ther. 1998; 286:447–458. PMID: 9655890.11. Zhang H, Cook J, Nickel J, Yu R, Stecker K, Myers K, Dean NM. Reduction of liver Fas expression by an antisense oligonucleotide protects mice from fulminant hepatitis. Nature Biotechnol. 2000; 18:862–867. PMID: 10932156.
Article12. Tomita N, Morishita R, Higaki J, Aoki M, Nakamura Y, Mikami H, Fukamizu A, Murakami K, Kaneda Y, Ogihara T. Transient decrease in high blood pressure by in vivo transfer of antisense oligodeoxynucleotides against rat angiotensinogen. Hypertension. 1995; 26:131–136. PMID: 7541778.13. Bennett CF, Cowsert LM. Antisense oligonucleotides as a tool for gene functionalization and target validation. Biochim Biophys Acta. 1999; 1489:19–30. PMID: 10806994.
Article14. Flanagan WM, Wagner RW. Potent and selective gene inhibition using antisense oligodeoxynucleotides. Mol Cell Biochem. 1997; 172:213–225. PMID: 9278247.
Article15. Matsuda M, Park JG, Wang DC, Hunter S, Chien P, Schreiber AD. Abrogation of the Fc gamma receptor IIA-mediated phagocytic signal by stem-loop Syk antisense oligonucleotides. Mol Biol Cell. 1996; 7:1095–1106. PMID: 8862523.16. Vickers TA, Wyatt JR, Freier SM. Effects of RNA secondary structure on cellular antisense activity. Nucl Acids Res. 2000; 28:1340–1347. PMID: 10684928.
Article17. Smith LK, Andersen B, Hovgaard L, Jaroszewski JW. Rational selection of antisense oligonucleotide sequences. Eur J Pharm Sci. 2000; 11:191–198. PMID: 11042224.
Article18. Moon IJ, Lee Y, Kwak CS, Lee JH, Choi K, Schreiber AD, Park JG. Target site search and effective inhibition of leukaemic cell growth by a covalently multiple antisense oligonucleotide to c-myb. Biochem J. 2000; 346:295–303. PMID: 10677346.19. Moon IJ, Choi K, Choi YK, Kim JE, Lee Y, Schreiber AD, Park JG. Potent growth inhibition of leukemic cells by novel ribbon-type antisense oligonucleotides to c-myb. J Biol Chem. 2000; 275:4647–4653. PMID: 10671493.20. Mann JR, Kasthuri N, Raafat F, Pincott JR, Parkes SE, Muir KR, Ingram LC, Cameron AH. Malignant hepatic tumours in children: Incidence, clinical features and aetiology. Paediatr Perinat Epidemiol. 1990; 4:276–289. PMID: 2374747.
Article21. Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, Lukyanov S, Lukyanov K, Gurskaya N, Sverdlov ED, Siebert PD. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci. 1996; 93:6025–6030. PMID: 8650213.
Article22. Sambrook J, Fritsch EF, Maniatis T, editors. Molecular cloning. 1989.23. Beuken E, Vink C, Bruggeman CA. One-step procedure for screening recombinant plasmids by size. Biotechniques. 1998; 24:748–750. PMID: 9591120.
Article24. Yonekura H, Migita H, Sakurai S, Wang H, Harada S, Abedin MJ, Yamagishi S, Yamamoto H. Antisense display, a method for functional gene screening: evaluation in a cell-free system and isolation of angiogenesis-related genes. Nucleic Acids Res. 1999; 27:2591–2600. PMID: 10373574.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Ovarian cancer related gene targeting with large circular antisense library
- Screening of Cell Cycle-Related Genes of Pleurotus eryngii Using Yeast Mutant Strains
- The effect of YB-1 antisense oligonucleotides on tumor cell growth
- Genomic Profiling of Liver Cancer
- Development of Covalently Closed c-myb Antisense Oligonucleotides for Growth Inhibition of Leukemic Cells