J Bacteriol Virol.
2006 Sep;36(3):185-194. 10.4167/jbv.2006.36.3.185.
Molecular Characterization of Porcine Endogenous Retrovirus gag Genes from Pigs in Korea
- Affiliations
-
- 1Department of Biotechnology, College of Animal Bioscience & Technology, Konkuk University, Korea. Kimera@konkuk.ac.kr
- 2Department of Animal Resource Science, College of Industrial Science, Kongju National University, Korea.
- KMID: 2055026
- DOI: http://doi.org/10.4167/jbv.2006.36.3.185
Abstract
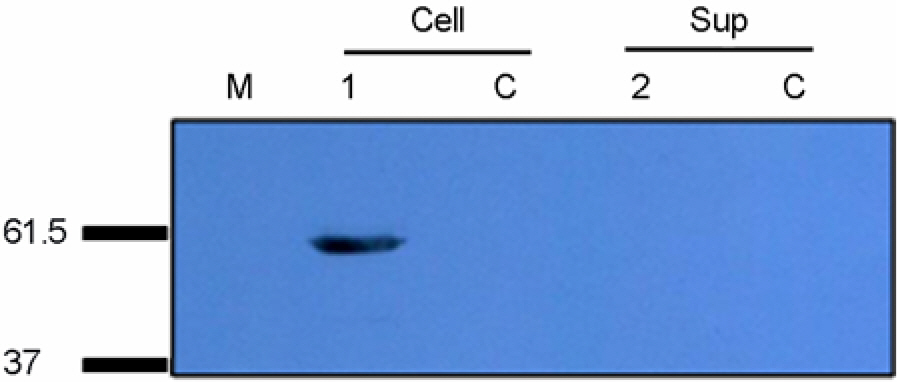
- Xenotransplantation, as a potential solution to the shortage of human organs, is associated with a number of concerns including immunologic rejection and xenogenic infection. While the pigs are considered the most suitable organ source for xenotransplantation, there is a potential public health risk due to zoonosis. Among the known porcine zoonotic microbes, Porcine Endogenous Retrovirus (PERV) is the most considerable virus. PERV belongs to the Gammaretrovirus and has been divided into three groups (A, B, and C). To characterize the gag of PERVs, we isolated the genomic DNAs from three pig breeds (Birkshire, Duroc, and Yorkshire) and two types of SPF miniature pigs. About 1.5 kb fragments covering full length of gag were amplified and cloned into T-vector. A total of 38 clones were obtained and sequenced. Nucleotide sequences were analyzed and phylogenetic trees were constructed from the nucleotide and deduced amino acids. PERV-A, -B and -C were present in the proportion of 47, 19 and 34%, respectively. Regardless of origin or subgroups, gag clones showed highly homology in nucleotide and deduced amino acid sequences. Deduced amino acids sequence alignments showed typical conserve sequences, Cys-His box and processing sites. Among analyzed clones, about 28% of isolates had the correct open reading frame. To test the functional expression of Gag protein, gag was subcloned into expression vector and confirmed its expression in HeLa cell. This research provides the fundamental information about molecular characteristics of gag gene and functional Gag protein related xenotropic PERVs.
Keyword
MeSH Terms
Figure
Reference
-
References
1). Akiyoshi DE, Denaro M, Zhu H, Greenstein JL, Banerjee P, Fishman JA. Identification of a full-length cDNA for an endogenous retrovirus of miniature swine. J Virol. 72:4503–4507. 1998.
Article2). Alin K, Goff SP. Amino acid substitutions in the CA protein of Moloney murine leukemia virus that block early events in infection. Virology. 222:339–351. 1996.
Article3). Blusch JH, Patience C, Martin U. Pig endogenous retro-viruses and xenotransplantation. Xenotransplantation. 9:242–251. 2002.
Article4). Breese SS Jr. Virus-like particles occurring in cultures of stable pig kidney cell lines. Brief Report Arch Gesamte Virusforsch. 30:401–404. 1970.
Article5). Burland TG. DNASTAR's Lasergene sequence analysis software. Methods Mol Biol. 131:71–91. 2000.
Article6). Cairns TM, Craven RC. Viral DNA synthesis defects in assembly-competent Rous sarcoma virus CA mutants. J Virol. 75:242–250. 2001.
Article7). Cannon PM, Matthews S, Clark N, Byles ED, Iourin O, Hockley DJ, Kingsman SM, Kingsman AJ. Structure-function studies of the human immunodeficiency virus type 1 matrix protein, p17. J Virol. 71:3474–3483. 1997.
Article8). Choi G, Park S, Choi B, Hong S, Lee J, Hunter E, Rhee SS. Identification of a cytoplasmic targeting/retention signal in a retroviral Gag polyprotein. J Virol. 73:5431–5437. 1999.
Article9). Craven RC, Leure-du Pree AE, Weldon RA Jr, Wills JW. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 69:4213–4227. 1995.
Article10). Crawford S, Goff SP. Mutations in gag proteins P12 and P15 of Moloney murine leukemia virus block early stages of infection. J Virol. 49:909–917. 1984.
Article11). D'Souza V, Summers MF. How retroviruses select their genomes. Nat Rev Microbiol. 3:643–655. 2005.12). Hanger JJ, Bromham LD, McKee JJ, O'Brien TM, Robinson WF. The nucleotide sequence of koala (Phascolarctos cinereus) retrovirus: a novel type C endogenous virus related to Gibbon ape leukemia virus. J Virol. 74:4264–4272. 2000.13). Heneine W, Tibell A, Switzer WM, Sandstrom P, Rosales GV, Mathews A, Korsgren O, Chapman E, Folks TM, Groth CG. No evidence of infection with porcine endogenous retrovirus in recipients of porcine islet-cell xenografts. Lancet. 352:695–699. 1998.
Article14). Hall BG. Comparison of the accuracies of several phylogenetic methods using protein and DNA sequences. Mol Biol Evol. 22:792–802. 2005.
Article15). Jorgensen EC, Pedersen FS, Jorgensen P. Matrix protein of Akv murine leukemia virus: genetic mapping of regions essential for particle formation. J Virol. 66:4479–4487. 1992.
Article16). Blusch Jürgen H., Sigrid Seelmeir, Klaus von der Helm. Molecular and Enzymatic haracterization of the Porcine Endogenous Retrovirus Protease. J Virol. 76:7913–7917. 2002.17). Young Kelly R., Ross Ted M.Elicitation of Immunity to HIV Type 1 Gag Is Determined by Gag Structure. AIDS RESEARCH AND HUMAN RETROVIRUSES. 22:99–108. 2006.
Article18). Lee EG, Alidina A, May C, Linial ML. Importance of basic residues in binding of rous sarcoma virus nucleocapsid to the RNA packaging signal. J Virol. 77:2010–2020. 2003.
Article19). Lee SK, Nagashima K, Hu WS. Cooperative effect of gag proteins p12 and capsid during early events of murine leukemia virus replication. J Virol. 79:4159–4169. 2005.
Article20). Le Tissier P, Stoye JP, Takeuchi Y, Patience C, Weiss RA. Two sets of human-tropic pig retrovirus. Nature. 389:681–682. 1997.
Article21). Magre S, Takeuchi Y, Bartosch B. Xenotransplantation and pig endogenous retroviruses. Rev Med Virol. 13:311–329. 2003.
Article22). Martina Y, Marcucci KT, Cherqui S, Szabo A, Drysdale T, Srinivisan U, Wilson CA, Patience C, Salomon DR. Mice transgenic for a human porcine endogenous retrovirus receptor are susceptible to productive viral infection. J Virol. 80:3135–3146. 2006.
Article23). Meric C, Goff SP. Characterization of Moloney murine leukemia virus mutants with single-amino-acid substitutions in the Cys-His box of the nucleocapsid protein. J Viro. 63:1558–1568. 1989.24). Patience C, Patton GS, Takeuchi Y, Weiss RA, McClure MO, Rydberg L, Breimer ME. No evidence of pig DNA or retroviral infection in patients with short-term extracorporeal connection to pig kidneys. Lancet. 352:699–701. 1998.
Article25). Schwartzberg P, Colicelli J, Gordon ML, Goff SP. Mutations in the gag gene of Moloney murine leukemia virus: effects on production of virions and reverse transcriptase. J Virol. 49:918–924. 1984.
Article26). Todaro GJ, Benveniste RE, Lieber MM, Sherr CJ. Characterization of a type C virus released from the porcine cell line PK(15). Virology. 58:65–74. 1974.
Article27). Tristem M, Kabat P, Lieberman L, Linde S, Karpas A, Hill F. Characterization of a novel murine leukemia virus-related subgroup within mammals. J Virol. 70:8241–8246. 1996.
Article28). Van de Peer Y, De Wachter R. Construction of evolutionary distance trees with TREECON for Windows: accounting for variation in nucleotide substitution rate among sites. Comput Appl Biosci. 13:227–230. 1997.
Article29). Yamashita M, Emerman M. Capsid is a dominant determinant of retrovirus infectivity in nondividing cells. J Virol. 78:5670–5678. 2004.
Article30). Zhang Y, Barklis E. Nucleocapsid protein effects on the specificity of retrovirus RNA encapsidation. J Virol. 69:5716–5722. 1995.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Analysis of swine leukocyte antigen class I gene profiles and porcine endogenous retrovirus viremia level in a transgenic porcine herd inbred for xenotransplantation research
- Analysis of env Subtypes of Porcine Endogenous Retrovirus in SNU Miniature Pigs
- No expression of porcine endogenous retrovirus after pig to monkey xenotransplantation
- Expression and Packaging of a Human Endogenous Retrovirus-K Genomic DNA Clone
- Comparison of the age-related porcine endogenous retrovirus (PERV) expression using duplex RT-PCR