Yonsei Med J.
2014 Sep;55(5):1206-1213. 10.3349/ymj.2014.55.5.1206.
Overexpression of Mucin 13 due to Promoter Methylation Promotes Aggressive Behavior in Ovarian Cancer Cells
- Affiliations
-
- 1Department of Biochemistry, School of Medicine, Ewha Womans University, Seoul, Korea. ahnj@ewha.ac.kr
- 2College of Pharmacy, Sunchon National University, Suncheon, Korea.
- 3Department of Obstetrics and Gynecology, School of Medicine, Ewha Womans University, Seoul, Korea. goodmorning@ewha.ac.kr
- KMID: 1799482
- DOI: http://doi.org/10.3349/ymj.2014.55.5.1206
Abstract
- PURPOSE
Recent discoveries suggest that aberrant DNA methylation provides cancer cells with advanced metastatic properties. However, the precise regulatory mechanisms controlling metastasis genes and their role in metastatic transformation are largely unknown. To address epigenetically-regulated gene products involved in ovarian cancer metastasis, we examined the mechanisms regulating mucin 13 (MUC13) expression and its influence on aggressive behaviors of ovarian malignancies.
MATERIALS AND METHODS
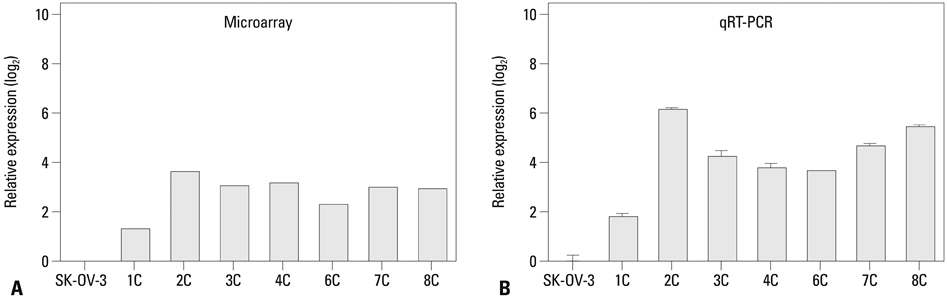
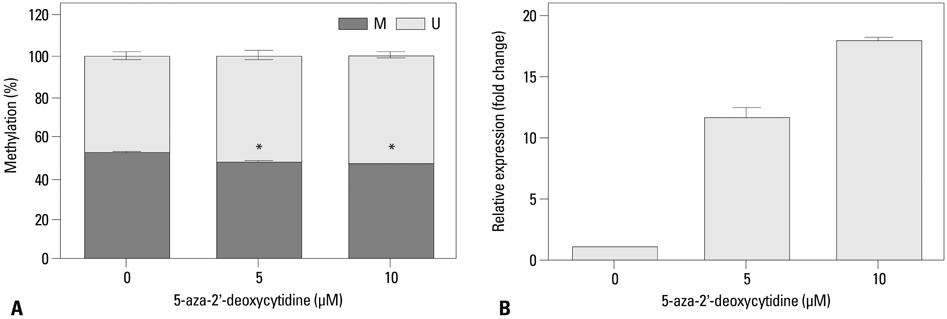
We injected SK-OV-3 ovarian cancer cells peritoneally into nude mice to mimic human ovarian tumor metastasis. Overexpression of MUC13 mRNA was detected in metastatic implants from the xenografts by expression microarray analysis and quantitative reverse-transcription polymerase chain reaction (qRT-PCR). The DNA methylation status within the MUC13 promoter region was determined using bisulfite sequencing PCR and quantitative methylation-specific PCR. We evaluated the effects of exogenous MUC13 on cell invasion and migration using in vitro transwell assays.
RESULTS
MUC13 mRNA expression was up-regulated, and methylation of specific CpG sites within the promoter was reduced in the metastatic implants relative to those in wild-type SK-OV-3 cells. Addition of a DNA methyltransferase inhibitor to SK-OV-3 cells induced MUC13 expression, thereby implying epigenetic regulation of MUC13 by promoter methylation. MUC13 overexpression increased migration and invasiveness, compared to control cells, suggesting aberrant up-regulation of MUC13 is strongly associated with progression of aggressive behaviors in ovarian cancer.
CONCLUSION
We provide novel evidence for epigenetic regulation of MUC13 in ovarian cancer. We suggest that the DNA methylation status within the MUC13 promoter region may be a potential biomarker of aggressive behavior in ovarian cancer.
Keyword
MeSH Terms
-
Animals
Cell Line, Tumor
*DNA Methylation
Epigenesis, Genetic
Female
*Gene Expression Regulation, Neoplastic
Heterografts/metabolism
Humans
Mice
Mice, Nude
Mucins/*genetics/metabolism/physiology
Neoplasm Invasiveness/genetics
Ovarian Neoplasms/genetics/*metabolism/pathology
RNA, Messenger/metabolism
Mucins
RNA, Messenger
Figure
Cited by 2 articles
-
Aberrant Hypomethylation of Solute Carrier Family 6 Member 12 Promoter Induces Metastasis of Ovarian Cancer
Hye Youn Sung, San-Duk Yang, Ae Kyung Park, Woong Ju, Jung-Hyuck Ahn
Yonsei Med J. 2017;58(1):27-34. doi: 10.3349/ymj.2017.58.1.27.Ovarian Clear Cell Carcinoma Sub-Typing by ARID1A Expression
Jae Yoon Choi, Hyun Ho Han, Young Tae Kim, Joo Hyun Lee, Baek Gil Kim, Suki Kang, Nam Hoon Cho
Yonsei Med J. 2017;58(1):59-66. doi: 10.3349/ymj.2017.58.1.59.
Reference
-
1. Lengyel E. Ovarian cancer development and metastasis. Am J Pathol. 2010; 177:1053–1064.
Article2. Kipps E, Tan DS, Kaye SB. Meeting the challenge of ascites in ovarian cancer: new avenues for therapy and research. Nat Rev Cancer. 2013; 13:273–282.
Article3. Rose MC, Voynow JA. Respiratory tract mucin genes and mucin glycoproteins in health and disease. Physiol Rev. 2006; 86:245–278.
Article4. Singh AP, Senapati S, Ponnusamy MP, Jain M, Lele SM, Davis JS, et al. Clinical potential of mucins in diagnosis, prognosis, and therapy of ovarian cancer. Lancet Oncol. 2008; 9:1076–1085.
Article5. Hollingsworth MA, Swanson BJ. Mucins in cancer: protection and control of the cell surface. Nat Rev Cancer. 2004; 4:45–60.
Article6. Gupta BK, Maher DM, Ebeling MC, Sundram V, Koch MD, Lynch DW, et al. Increased expression and aberrant localization of mucin 13 in metastatic colon cancer. J Histochem Cytochem. 2012; 60:822–831.
Article7. Ringel J, Löhr M. The MUC gene family: their role in diagnosis and early detection of pancreatic cancer. Mol Cancer. 2003; 2:9.8. Brockhausen I, Yang JM, Burchell J, Whitehouse C, Taylor-Papadimitriou J. Mechanisms underlying aberrant glycosylation of MUC1 mucin in breast cancer cells. Eur J Biochem. 1995; 233:607–617.
Article9. Rakha EA, Boyce RW, Abd El-Rehim D, Kurien T, Green AR, Paish EC, et al. Expression of mucins (MUC1, MUC2, MUC3, MUC4, MUC5AC and MUC6) and their prognostic significance in human breast cancer. Mod Pathol. 2005; 18:1295–1304.
Article10. Rosen DG, Wang L, Atkinson JN, Yu Y, Lu KH, Diamandis EP, et al. Potential markers that complement expression of CA125 in epithelial ovarian cancer. Gynecol Oncol. 2005; 99:267–277.
Article11. Dong Y, Walsh MD, Cummings MC, Wright RG, Khoo SK, Parsons PG, et al. Expression of MUC1 and MUC2 mucins in epithelial ovarian tumours. J Pathol. 1997; 183:311–317.
Article12. Giuntoli RL 2nd, Rodriguez GC, Whitaker RS, Dodge R, Voynow JA. Mucin gene expression in ovarian cancers. Cancer Res. 1998; 58:5546–5550.13. Chauhan SC, Vannatta K, Ebeling MC, Vinayek N, Watanabe A, Pandey KK, et al. Expression and functions of transmembrane mucin MUC13 in ovarian cancer. Cancer Res. 2009; 69:765–774.
Article14. Canney PA, Moore M, Wilkinson PM, James RD. Ovarian cancer antigen CA125: a prospective clinical assessment of its role as a tumour marker. Br J Cancer. 1984; 50:765–769.
Article15. Yin BW, Dnistrian A, Lloyd KO. Ovarian cancer antigen CA125 is encoded by the MUC16 mucin gene. Int J Cancer. 2002; 98:737–740.16. Budiu RA, Mantia-Smaldone G, Elishaev E, Chu T, Thaller J, McCabe K, et al. Soluble MUC1 and serum MUC1-specific antibodies are potential prognostic biomarkers for platinum-resistant ovarian cancer. Cancer Immunol Immunother. 2011; 60:975–984.
Article17. Chauhan SC, Singh AP, Ruiz F, Johansson SL, Jain M, Smith LM, et al. Aberrant expression of MUC4 in ovarian carcinoma: diagnostic significance alone and in combination with MUC1 and MUC16 (CA125). Mod Pathol. 2006; 19:1386–1394.
Article18. Williams SJ, Wreschner DH, Tran M, Eyre HJ, Sutherland GR, McGuckin MA. Muc13, a novel human cell surface mucin expressed by epithelial and hemopoietic cells. J Biol Chem. 2001; 276:18327–18336.
Article19. Maher DM, Gupta BK, Nagata S, Jaggi M, Chauhan SC. Mucin 13: structure, function, and potential roles in cancer pathogenesis. Mol Cancer Res. 2011; 9:531–537.
Article20. Walsh MD, Young JP, Leggett BA, Williams SH, Jass JR, McGuckin MA. The MUC13 cell surface mucin is highly expressed by human colorectal carcinomas. Hum Pathol. 2007; 38:883–892.
Article21. Shimamura T, Ito H, Shibahara J, Watanabe A, Hippo Y, Taniguchi H, et al. Overexpression of MUC13 is associated with intestinal-type gastric cancer. Cancer Sci. 2005; 96:265–273.
Article22. Chauhan SC, Ebeling MC, Maher DM, Koch MD, Watanabe A, Aburatani H, et al. MUC13 mucin augments pancreatic tumorigenesis. Mol Cancer Ther. 2012; 11:24–33.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- DNA Hypomethylation-Mediated Overexpression of Carbonic Anhydrase 9 Induces an Aggressive Phenotype in Ovarian Cancer Cells
- Regulation of MUC6 Methylation Correlates with Progression of Gastric Cancer
- DNA methylation of Bcl-2 family genes in cancer cells
- Peripheral blood BRCA1 methylation profiling to predict familial ovarian cancer
- Aberrant epigenetic regulation of GABRP associates with aggressive phenotype of ovarian cancer