Ann Lab Med.
2020 Jan;40(1):63-67. 10.3343/alm.2020.40.1.63.
Comparison of 16S Ribosomal RNA Targeted Sequencing and Culture for Bacterial Identification in Normally Sterile Body Fluid Samples: Report of a 10-Year Clinical Laboratory Review
- Affiliations
-
- 1Department of Laboratory Medicine and Genetics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea. pmhhj77@gmail.com, micro.lee@samsung.com
- 2Division of Infectious Diseases, Department of Pediatrics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 3Division of Infectious Diseases, Department of Medicine, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- 4Center for Infection Prevention and Control, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea.
- KMID: 2457495
- DOI: http://doi.org/10.3343/alm.2020.40.1.63
Abstract
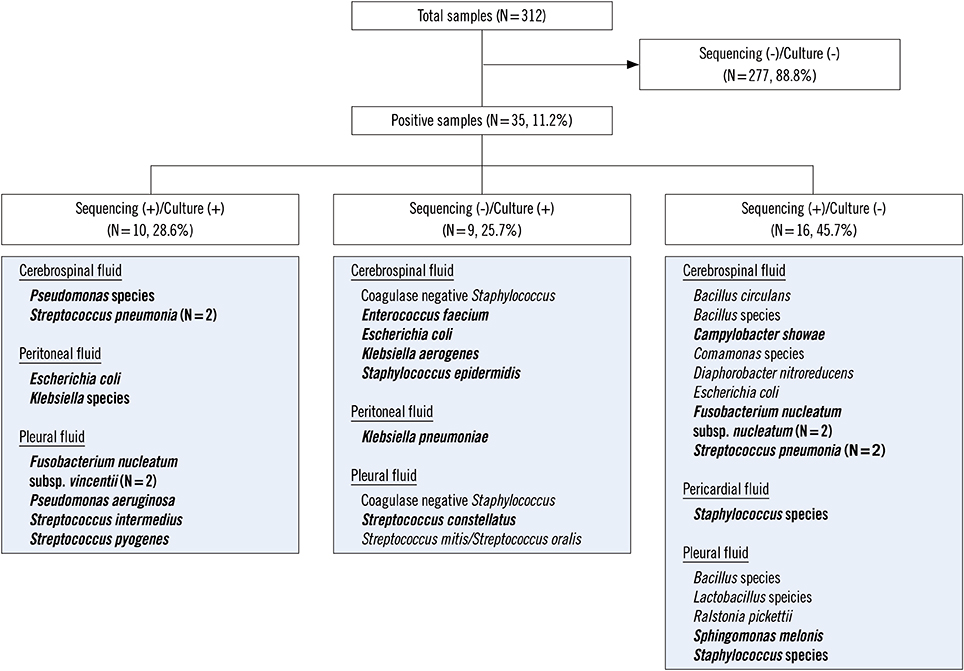
- As 16S ribosomal RNA (rRNA)-targeted sequencing can detect DNA from non-viable bacteria, it can be used to identify pathogens from clinical samples even in patients pretreated with antibiotics. We compared the results of 16S rRNA-targeted sequencing and culture for identifying bacterial species in normally sterile body fluid (NSBF): cerebrospinal, pericardial, peritoneal and pleural fluids. Over a 10-year period, a total of 312 NSBF samples were evaluated simultaneously using 16S rRNA-targeted sequencing and culture. Results were concordant in 287/312 (92.0%) samples, including 277 (88.8%) negative and 10 (3.2%) positive samples. Of the 16 sequencing-positive, culture-negative samples, eight showed clinically relevant isolates that included Fusobacterium nucleatum subsp. nucleatum, Streptococcus pneumoniae, and Staphylococcus spp. All these samples were obtained from the patients pretreated with antibiotics. The diagnostic yield of 16S rRNA-targeted sequencing combined with culture was 11.2%, while that of culture alone was 6.1%. 16S rRNA-targeted sequencing in conjunction with culture could be useful for identifying bacteria in NSBF samples, especially when patients have been pretreated with antibiotics and when anaerobic infection is suspected.
MeSH Terms
Figure
Reference
-
1. Altun O, Almuhayawi M, Ullberg M, Özenci V. Rapid identification of microorganisms from sterile body fluids by use of FilmArray. J Clin Microbiol. 2015; 53:710–712.
Article2. Grif K, Heller I, Prodinger WM, Lechleitner K, Lass-Flörl C, Orth D. Improvement of detection of bacterial pathogens in normally sterile body sites with a focus on orthopedic samples by use of a commercial 16S rRNA broad-range PCR and sequence analysis. J Clin Microbiol. 2012; 50:2250–2254.
Article3. Rampini SK, Bloemberg GV, Keller PM, Büchler AC, Dollenmaier G, Speck RF, et al. Broad-range 16S rRNA gene polymerase chain reaction for diagnosis of culture-negative bacterial infections. Clin Infect Dis. 2011; 53:1245–1251.
Article4. Sontakke S, Cadenas MB, Maggi RG, Diniz PP, Breitschwerdt EB. Use of broad range 16S rDNA PCR in clinical microbiology. J Microbiol Methods. 2009; 76:217–225.5. Leber AL, editor. Clinical microbiology procedures handbook. 4th ed. Washington, DC: ASM Press;2016. p. 3.5–3.7.6. CLSI. Interpretive criteria for identification of bacteria and fungi by targeted DNA sequencing. 2nd ed. CLSI MM18. Wayne, PA: Clinical and Laboratory Standards Institute;2018.7. Li X, Xing J, Li B, Wang P, Liu J. Use of tuf as a target for sequence-based identification of Gram-positive cocci of the genus Enterococcus, Streptococcus, coagulase-negative Staphylococcus, and Lactococcus. Ann Clin Microbiol Antimicrob. 2012; 11:31.8. Dauga C. Evolution of the gyrB gene and the molecular phylogeny of Enterobacteriaceae: a model molecule for molecular systematic studies. Int J Syst Evol Microbiol. 2002; 52:531–547.9. Basein T, Gardiner BJ, Andujar Vazquez GM, Joel Chandranesan AS, Rabson AR, Doron S, et al. Microbial identification using DNA target amplification and sequencing: clinical utility and impact on patient management. Open Forum Infect Dis. 2018; 5:ofy257.
Article10. Varani S, Stanzani M, Paolucci M, Melchionda F, Castellani G, Nardi L, et al. Diagnosis of bloodstream infections in immunocompromised patients by real-time PCR. J Infect. 2009; 58:346–351.
Article11. Welinder-Olsson C, Dotevall L, Hogevik H, Jungnelius R, Trollfors B, Wahl M, et al. Comparison of broad-range bacterial PCR and culture of cerebrospinal fluid for diagnosis of community-acquired bacterial meningitis. Clin Microbiol Infect. 2007; 13:879–886.
Article12. Fiore AE, Moroney JF, Farley MM, Harrison LH, Patterson JE, Jorgensen JH, et al. Clinical outcomes of meningitis caused by Streptococcus pneumoniae in the era of antibiotic resistance. Clin Infect Dis. 2000; 30:71–77.13. Kanegaye JT, Soliemanzadeh P, Bradley JS. Lumbar puncture in pediatric bacterial meningitis: defining the time interval for recovery of cerebrospinal fluid pathogens after parenteral antibiotic pretreatment. Pediatrics. 2001; 108:1169–1174.
Article14. Yang CC, Ye JJ, Hsu PC, Chang HJ, Cheng CW, Leu HS, et al. Characteristics and outcomes of Fusobacterium nucleatum bacteremia–a 6-year experience at a tertiary care hospital in northern Taiwan. Diagn Microbiol Infect Dis. 2011; 70:167–174.15. de Vries JJ, Arents NL, Manson WL. Campylobacter species isolated from extra-oro-intestinal abscesses: a report of four cases and literature review. Eur J Clin Microbiol Infect Dis. 2008; 27:1119–1123.16. Jenkins C, Ling CL, Ciesielczuk HL, Lockwood J, Hopkins S, McHugh TD, et al. Detection and identification of bacteria in clinical samples by 16S rRNA gene sequencing: comparison of two different approaches in clinical practice. J Med Microbiol. 2012; 61:483–488.
Article17. Velásquez-Mejía EP, de la Cuesta-Zuluaga J, Escobar JS. Impact of DNA extraction, sample dilution, and reagent contamination on 16S rRNA gene sequencing of human feces. Appl Microbiol Biotechnol. 2018; 102:403–411.
Article18. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014; 12:87.
Article19. Kolbert CP, Persing DH. Ribosomal DNA sequencing as a tool for identification of bacterial pathogens. Curr Opin Microbiol. 1999; 2:299–305.
Article20. Bourbeau P, Riley J, Heiter BJ, Master R, Young C, Pierson C. Use of the BacT/Alert blood culture system for culture of sterile body fluids other than blood. J Clin Microbiol. 1998; 36:3273–3277.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Three Cases of Moraxella osloensis Meningitis: A Difficult Experience in Species Identification and Determination of Clinical Significance
- Native Valve Endocarditis due to Corynebacterium striatum confirmed by 16S Ribosomal RNA Sequencing: A Case Report and Literature Review
- Laboratory Identification of Leptotrichia Species Isolated From Bacteremia Patients at a Single Institution
- Molecular Evolution and Identification of Yersinia Species by 16S rDNA Analysis
- Clinical and Microbiological Characteristics of Six Staphylococcus pettenkoferi Isolates From Blood Samples