Brain Tumor Res Treat.
2018 Apr;6(1):31-38. 10.14791/btrt.2018.6.e5.
Mitochondrial 10398A>G NADH-Dehydrogenase Subunit 3 of Complex I Is Frequently Altered in Intra-Axial Brain Tumors in Malaysia
- Affiliations
-
- 1Department of Neurosciences, School of Medical Sciences, Universiti Sains Malaysia, Health Campus, Kelantan, Malaysia. azizmdy@yahoo.com
- 2School of Health Sciences, Universiti Sains Malaysia, Health Campus, Kelantan, Malaysia.
- 3Center for Neuroscience Services and Research, Universiti Sains Malaysia, Health Campus, Kelantan, Malaysia.
- KMID: 2410235
- DOI: http://doi.org/10.14791/btrt.2018.6.e5
Abstract
- BACKGROUND
Mitochondria are major cellular sources of reactive oxygen species (ROS) generation which can induce mitochondrial DNA damage and lead to carcinogenesis. The mitochondrial 10398A>G alteration in NADH-dehydrogenase subunit 3 (ND3) can severely impair complex I, a key component of ROS production in the mitochondrial electron transport chain. Alteration in ND3 10398A>G has been reported to be linked with diverse neurodegenerative disorders and cancers. The aim of this study was to find out the association of mitochondrial ND3 10398A>G alteration in brain tumor of Malaysian patients.
METHODS
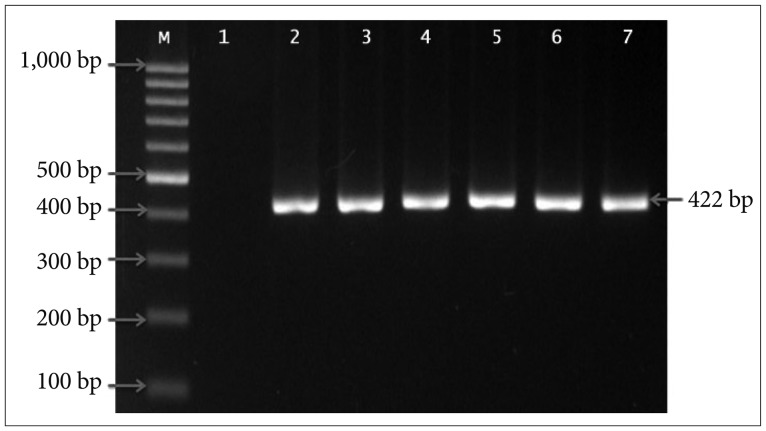
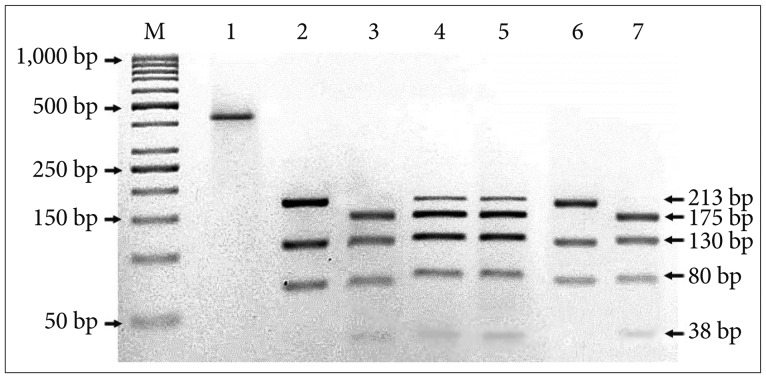
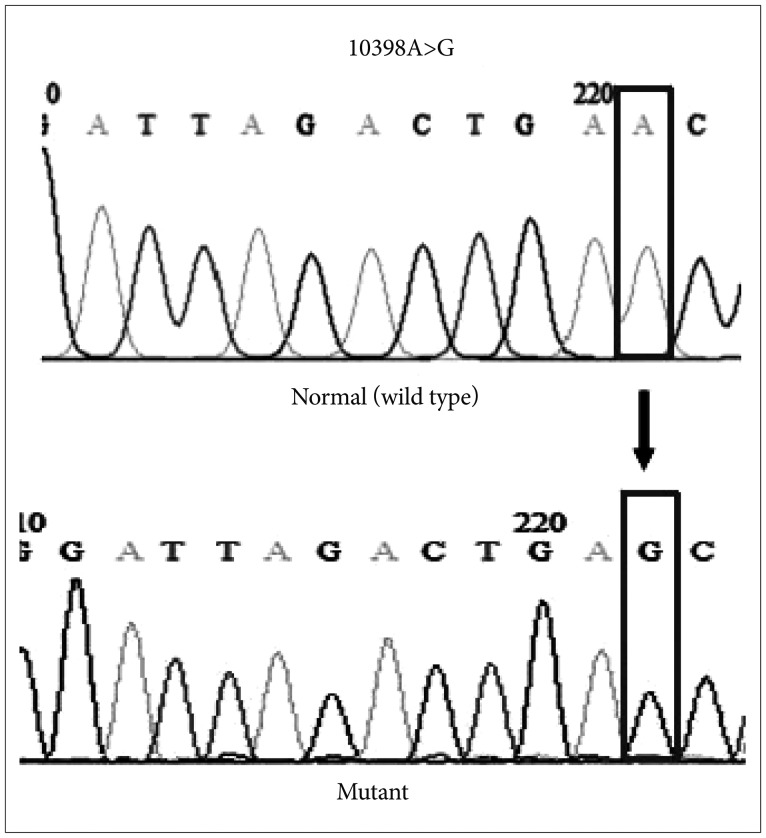
Brain tumor tissues and corresponding blood specimens were obtained from 45 patients. The ND3 10398A>G alteration at target codon 114 was detected using the PCR-RFLP analysis and later was confirmed by DNA sequencing.
RESULTS
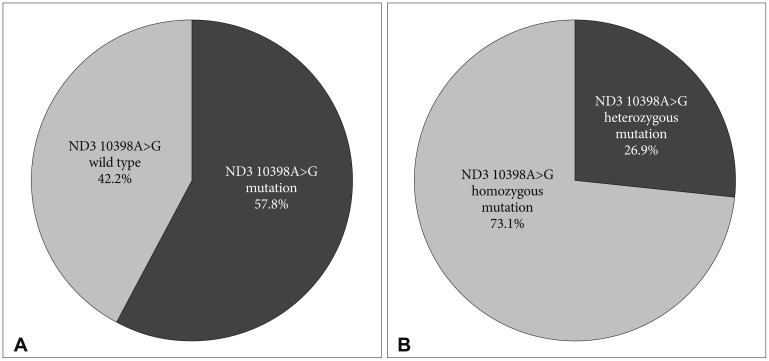
Twenty-six (57.8%) patients showed ND3 10398A>G mutation in their tumor specimens, in which 26.9% of these mutations were heterozygous mutations. ND3 10398A>G mutation was not significantly correlated with age, gender, and histological tumor grade, however was found more frequently in intra-axial than in extra-axial tumors (62.5% vs. 46.2%, p < 0.01).
CONCLUSION
For the first time, we have been able to describe the occurrence of ND3 10398A>G mutations in a Malaysian brain tumor population. It can be concluded that mitochondrial ND3 10398A>G alteration is frequently present in brain tumors among Malaysian population and it shows an impact on the intra-axial tumors.
Keyword
MeSH Terms
Figure
Reference
-
1. de Robles P, Fiest KM, Frolkis AD, et al. The worldwide incidence and prevalence of primary brain tumors: a systematic review and meta-analysis. Neuro Oncol. 2015; 17:776–783. PMID: 25313193.
Article2. National Brain Tumor Society. Quick Brain Tumor Facts. Newton, MA: National Brain Tumor Society;2018. Accessed April, 2018. http://braintumor.org/brain-tumor-information/brain-tumor-facts/.3. Lim GCC, Rampal S, Halimah Y. Cancer incidence in Peninsular Malaysia, 2003–2005. Kuala Lumpur: National Cancer Registry;2008.4. Goh CH, Lu YY, Lau BL, et al. Brain and spinal tumour. Med J Malaysia. 2014; 69:261–267. PMID: 25934956.5. Wallace DC, Fan W, Procaccio V. Mitochondrial energetics and therapeutics. Annu Rev Pathol. 2010; 5:297–348. PMID: 20078222.
Article6. Galluzzi L, Kepp O, Kroemer G. Mitochondria: master regulators of danger signalling. Nat Rev Mol Cell Biol. 2012; 13:780–788. PMID: 23175281.
Article7. Verschoor ML, Ungard R, Harbottle A, Jakupciak JP, Parr RL, Singh G. Mitochondria and cancer: past, present, and future. Biomed Res Int. 2013; 2013:612369. PMID: 23509753.
Article8. Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006; 25:4663–4674. PMID: 16892080.
Article9. Modica-Napolitano JS, Singh KK. Mitochondria as targets for detection and treatment of cancer. Expert Rev Mol Med. 2002; 4:1–19.
Article10. Modica-Napolitano JS, Singh KK. Mitochondrial dysfunction in cancer. Mitochondrion. 2004; 4:755–762. PMID: 16120430.
Article11. Singh KK, Kulawiec M. Mitochondrial DNA polymorphism and risk of cancer. Methods Mol Biol. 2009; 471:291–303. PMID: 19109786.
Article12. Chandra D, Singh KK. Genetic insights into OXPHOS defect and its role in cancer. Biochim Biophys Acta. 2011; 1807:620–625. PMID: 21074512.
Article13. Chatterjee A, Dasgupta S, Sidransky D. Mitochondrial subversion in cancer. Cancer Prev Res (Phila). 2011; 4:638–654. PMID: 21543342.
Article14. Tseng LM, Yin PH, Yang CW, et al. Somatic mutations of the mitochondrial genome in human breast cancers. Genes Chromosomes Cancer. 2011; 50:800–811. PMID: 21748819.
Article15. Hsu CC, Lee HC, Wei YH. Mitochondrial DNA alterations and mitochondrial dysfunction in the progression of hepatocellular carcinoma. World J Gastroenterol. 2013; 19:8880–8886. PMID: 24379611.
Article16. Yang Ai SS, Hsu K, Herbert C, et al. Mitochondrial DNA mutations in exhaled breath condensate of patients with lung cancer. Respir Med. 2013; 107:911–918. PMID: 23507584.
Article17. Li LH, Kang T, Chen L, et al. Detection of mitochondrial DNA mutations by high-throughput sequencing in the blood of breast cancer patients. Int J Mol Med. 2014; 33:77–82. PMID: 24253185.
Article18. Lee HC, Huang KH, Yeh TS, Chi CW. Somatic alterations in mitochondrial DNA and mitochondrial dysfunction in gastric cancer progression. World J Gastroenterol. 2014; 20:3950–3959. PMID: 24744584.
Article19. Fang Y, Huang J, Zhang J, et al. Detecting the somatic mutations spectrum of Chinese lung cancer by analyzing the whole mitochondrial DNA genomes. Mitochondrial DNA. 2015; 26:56–60. PMID: 24006865.
Article20. Hsu CC, Tseng LM, Lee HC. Role of mitochondrial dysfunction in cancer progression. Exp Biol Med (Maywood). 2016; 241:1281–1295. PMID: 27022139.
Article21. Canter JA, Kallianpur AR, Parl FF, Millikan RC. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2005; 65:8028–8033. PMID: 16140977.
Article22. Darvishi K, Sharma S, Bhat AK, Rai E, Bamezai RN. Mitochondrial DNA G10398A polymorphism imparts maternal Haplogroup N a risk for breast and esophageal cancer. Cancer Lett. 2007; 249:249–255. PMID: 17081685.
Article23. Kulawiec M, Owens KM, Singh KK. mtDNA G10398A variant in African-American women with breast cancer provides resistance to apoptosis and promotes metastasis in mice. J Hum Genet. 2009; 54:647–654. PMID: 19763141.
Article24. Jiang H, Zhao H, Xu H, et al. Peripheral blood mitochondrial DNA content, A10398G polymorphism, and risk of breast cancer in a Han Chinese population. Cancer Sci. 2014; 105:639–645. PMID: 24703408.25. van der Walt JM, Dementieva YA, Martin ER, et al. Analysis of European mitochondrial haplogroups with Alzheimer disease risk. Neurosci Lett. 2004; 365:28–32. PMID: 15234467.
Article26. Shoffner JM, Brown MD, Torroni A, et al. Mitochondrial DNA variants observed in Alzheimer disease and Parkinson disease patients. Genomics. 1993; 17:171–184. PMID: 8104867.
Article27. Kösel S, Grasbon-Frodl EM, Mautsch U, et al. Novel mutations of mitochondrial complex I in pathologically proven Parkinson disease. Neurogenetics. 1998; 1:197–204. PMID: 10737123.
Article28. van der Walt JM, Nicodemus KK, Martin ER, et al. Mitochondrial polymorphisms significantly reduce the risk of Parkinson disease. Am J Hum Genet. 2003; 72:804–811. PMID: 12618962.
Article29. Bai RK, Leal SM, Covarrubias D, Liu A, Wong LJ. Mitochondrial genetic background modifies breast cancer risk. Cancer Res. 2007; 67:4687–4694. PMID: 17510395.
Article30. Setiawan VW, Chu LH, John EM, et al. Mitochondrial DNA G10398A variant is not associated with breast cancer in African-American women. Cancer Genet Cytogenet. 2008; 181:16–19. PMID: 18262047.
Article31. Czarnecka AM, Krawczyk T, Zdrozny M, et al. Mitochondrial NADH-dehydrogenase subunit 3 (ND3) polymorphism (A10398G) and sporadic breast cancer in Poland. Breast Cancer Res Treat. 2010; 121:511–518. PMID: 19266278.
Article32. Mohamed Yusoff AA, Mohd Nasir KN, Haris K, et al. Detection of somatic mutations in the mitochondrial DNA control region D-loop in brain tumors: the first report in Malaysian patients. Oncol Lett. 2017; 14:5179–5188. PMID: 29098023.
Article33. Louis DN, Ohgaki H, Wiestler OD, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007; 114:97–109. PMID: 17618441.
Article34. Mohamed Yusoff AA, Zulfakhar FN, Sul'ain MD, Idris Z, Abdullah JM. Association of the IDH1 C.395G>A (R132H) mutation with histological type in Malay brain tumors. Asian Pac J Cancer Prev. 2016; 17:5195–5201. PMID: 28125199.35. Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992; 61:1175–1212. PMID: 1497308.
Article36. Dimauro S, Davidzon G. Mitochondrial DNA and disease. Ann Med. 2005; 37:222–232. PMID: 16019721.
Article38. Grzybowska-Szatkowska L, Slaska B. Mitochondrial DNA and carcinogenesis (review). Mol Med Rep. 2012; 6:923–930. PMID: 22895648.39. Mims MP, Hayes TG, Zheng S, et al. Mitochondrial DNA G10398A polymorphism and invasive breast cancer in African-American women. Cancer Res. 2006; 66:1880. author reply 1880-1. PMID: 16452251.
Article40. Sultana GNN, Rahman A, Karim MM, Shahinuzzaman ADA, Begum R, Begum RA. Breast cancer risk associated mitochondrial NADHdehydrogenase subunit-3 (ND3) polymorphisms (G10398A and T10400C) in Bangladeshi women. J Med Genet Genomics. 2011; 3:131–135.41. Tengku Baharudin N, Jaafar H, Zainuddin Z. Association of mitochondrial DNA 10398 polymorphism in invasive breast cancer in malay population of peninsular malaysia. Malays J Med Sci. 2012; 19:36–42.42. Yu Y, Lv F, Lin H, et al. Mitochondrial ND3 G10398A mutation: a biomarker for breast cancer. Genet Mol Res. 2015; 14:17426–17431. PMID: 26782384.
Article43. Ismaeel HM, Younan HQ, Zahid RA. Mitochondrial DNA A10398G mutation is not associated with breast cancer risk in a sample of Iraqi women. Curr Res J Biol Sci. 2013; 5:126–129.44. Yeh JJ, Lunetta KL, van Orsouw NJ, et al. Somatic mitochondrial DNA (mtDNA) mutations in papillary thyroid carcinomas and differential mtDNA sequence variants in cases with thyroid tumours. Oncogene. 2000; 19:2060–2066. PMID: 10803467.
Article45. Weller M, van den Bent M, Hopkins K, et al. EANO guideline for the diagnosis and treatment of anaplastic gliomas and glioblastoma. Lancet Oncol. 2014; 15:e395–e403. PMID: 25079102.
Article46. Wang XW, Ciccarino P, Rossetto M, et al. IDH mutations: genotype-phenotype correlation and prognostic impact. Biomed Res Int. 2014; 2014:540236. PMID: 24877111.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Effects of chronic alcohol and excessive iron intake on mitochondrial DNA damage in the rat liver
- Anesthetic experience in a pediatric patient with pyruvate dehydrogenase complex (PDHC) deficiency: A case report
- Molecular Detection of Spirometra decipiens in the United States
- A MELAS syndrome family harboring two mutations in mitochondrial genome
- A Novel Nicotinamide Adenine Dinucleotide Correction Method for Mitochondrial Ca2+ Measurement with FURA-2-FF in Single Permeabilized Ventricular Myocytes of Rat