Ann Lab Med.
2018 Mar;38(2):110-118. 10.3343/alm.2018.38.2.110.
Utility of Conventional Culture and MALDI-TOF MS for Identification of Microbial Communities in Bronchoalveolar Lavage Fluid in Comparison with the GS Junior Next Generation Sequencing System
- Affiliations
-
- 1Research Institute of Bacterial Resistance and Department of Laboratory Medicine, Yonsei University College of Medicine, Seoul, Korea. DEYONG@yuhs.ac
- 2Brain Korea 21 PLUS Project for Medical Science, Yonsei University, Seoul, Korea.
- 3Division of Pulmonary and Critical Care Medicine, Department of Internal Medicine, Severance Hospital, Yonsei University College of Medicine, Seoul, Korea.
- 4Department of Internal Medicine, Seoul National University College of Medicine, Division of Pulmonary and Critical Care Medicine, Seoul National University Bundang Hospital, Seongnam, Korea.
- KMID: 2403355
- DOI: http://doi.org/10.3343/alm.2018.38.2.110
Abstract
- BACKGROUND
Diverse microbiota exist in the lower respiratory tract. Although next generation sequencing (NGS) is the most widely used microbiome analysis technique, it is difficult to implement NGS in clinical microbiology laboratories. Therefore, we evaluated the performance of conventional culture methods together with matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) in identifying microbiota in bronchoalveolar lavage (BAL) fluid.
METHODS
BAL fluid samples (n=27) were obtained from patients undergoing diagnostic bronchoscopy for lung mass evaluation. Bacterial and fungal culture was performed with conventional media used in clinical microbiology laboratories. On an average, 20 isolated colonies were picked from each agar plate and identified by MALDI-TOF MS. Microbiome analysis using 16S rRNA NGS was conducted for comparison.
RESULTS
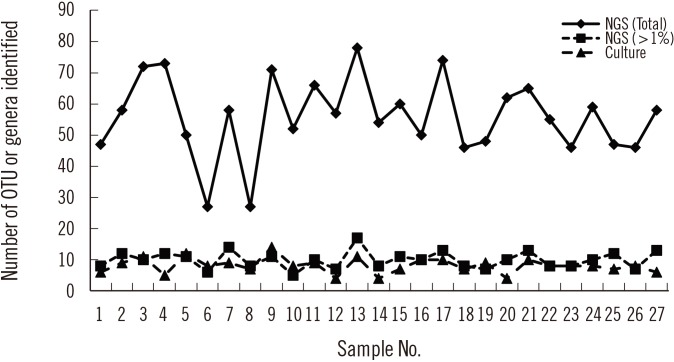
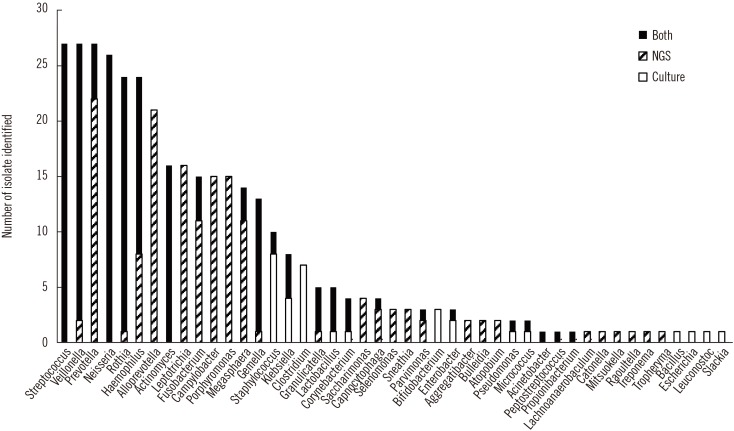
Streptococcus spp. and Neisseria spp. were most frequently cultured from the BAL fluid samples. In two samples, Enterobacteriaceae grew predominantly on MacConkey agar. Actinomyces and Veillonella spp. were commonly identified anaerobes; gut bacteria, such as Lactobacillus, Bifidobacterium, and Clostridium, and fungi were also isolated. NGS revealed more diverse bacterial communities than culture, and Prevotella spp. were mainly identified solely by NGS. Some bacteria, such as Staphylococcus spp., Clostridium spp., and Bifidobacterium spp., were identified solely by culture, indicating that culture may be more sensitive for detecting certain bacteria.
CONCLUSIONS
Culture and NGS of BAL fluid samples revealed common bacteria with some different microbial communities. Despite some limitations, culture combined with MALDI-TOF MS might play a complementary role in microbiome analysis using 16S rRNA NGS.
Keyword
MeSH Terms
-
Actinomyces
Agar
Bacteria
Bifidobacterium
Bronchoalveolar Lavage Fluid*
Bronchoalveolar Lavage*
Bronchoscopy
Clostridium
Enterobacteriaceae
Fungi
Humans
Lactobacillus
Lung
Mass Spectrometry
Microbiota
Neisseria
Prevotella
Respiratory System
Spectrometry, Mass, Matrix-Assisted Laser Desorption-Ionization
Staphylococcus
Streptococcus
Veillonella
Agar
Figure
Reference
-
1. Twigg HL 3rd, Morris A, Ghedin E, Curtis JL, Huffnagle GB, Crothers K, et al. Use of bronchoalveolar lavage to assess the respiratory microbiome: signal in the noise. Lancet Respir Med. 2013; 1:354–356. PMID: 24429191.2. Charlson ES, Bittinger K, Haas AR, Fitzgerald AS, Frank I, Yadav A, et al. Topographical continuity of bacterial populations in the healthy human respiratory tract. Am J Respir Crit Care Med. 2011; 184:957–963. PMID: 21680950.3. Erb-Downward JR, Thompson DL, Han MK, Freeman CM, McCloskey L, Schmidt LA, et al. Analysis of the lung microbiome in the “healthy” smoker and in COPD. PLoS One. 2011; 6:e16384. PMID: 21364979.4. Sze MA, Dimitriu PA, Hayashi S, Elliott WM, McDonough JE, Gosselink JV, et al. The lung tissue microbiome in chronic obstructive pulmonary disease. Am J Respir Crit Care Med. 2012; 185:1073–1080. PMID: 22427533.5. Morris A, Beck JM, Schloss PD, Campbell TB, Crothers K, Curtis JL, et al. Comparison of the respiratory microbiome in healthy nonsmokers and smokers. Am J Respir Crit Care Med. 2013; 187:1067–1075. PMID: 23491408.6. Hamady M, Knight R. Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Res. 2009; 19:1141–1152. PMID: 19383763.7. Allen-Vercoe E. Bringing the gut microbiota into focus through microbial culture: recent progress and future perspective. Curr Opin Microbiol. 2013; 16:625–629. PMID: 24148301.8. Sommer MO. Advancing gut microbiome research using cultivation. Curr Opin Microbiol. 2015; 27:127–132. PMID: 26401902.9. Goodman AL, Kallstrom G, Faith JJ, Reyes A, Moore A, Dantas G, et al. Extensive personal human gut microbiota culture collections characterized and manipulated in gnotobiotic mice. Proc Natl Acad Sci U S A. 2011; 108:6252–6257. PMID: 21436049.10. Lagier JC, Armougom F, Million M, Hugon P, Pagnier I, Robert C, et al. Microbial culturomics: paradigm shift in the human gut microbiome study. Clin Microbiol Infect. 2012; 18:1185–1193. PMID: 23033984.11. Rettedal EA, Gumpert H, Sommer MO. Cultivation-based multiplex phenotyping of human gut microbiota allows targeted recovery of previously uncultured bacteria. Nat Commun. 2014; 5:4714. PMID: 25163406.12. Brady C, Cleenwerck I, Venter S, Vancanneyt M, Swings J, Coutinho T. Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst Appl Microbiol. 2008; 31:447–460. PMID: 19008066.13. Heikens E, Fleer A, Paauw A, Florijn A, Fluit AC. Comparison of genotypic and phenotypic methods for species-level identification of clinical isolates of coagulase-negative staphylococci. J Clin Microbiol. 2005; 43:2286–2290. PMID: 15872257.14. Neuberger A, Oren I, Sprecher H. Clinical impact of a PCR assay for rapid identification of Klebsiella pneumoniae in blood cultures. J Clin Microbiol. 2008; 46:377–379. PMID: 17942663.15. Liu Z, Lozupone C, Hamady M, Bushman FD, Knight R. Short pyrosequencing reads suffice for accurate microbial community analysis. Nucleic Acids Res. 2007; 35:e120. PMID: 17881377.16. Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011; 27:2194–2200. PMID: 21700674.17. Kim OS, Cho YJ, Lee K, Yoon SH, Kim M, Na H, et al. Introducing EzTaxon-e: a prokaryotic 16S rRNA gene sequence database with phylotypes that represent uncultured species. Int J Syst Evol Microbiol. 2012; 62:716–721. PMID: 22140171.18. Charlson ES, Bittinger K, Chen J, Diamond JM, Li H, Collman RG, et al. Assessing bacterial populations in the lung by replicate analysis of samples from the upper and lower respiratory tracts. PLoS One. 2012; 7:e42786. PMID: 22970118.19. Bassis CM, Erb-Downward JR, Dickson RP, Freeman CM, Schmidt TM, Young VB, et al. Analysis of the upper respiratory tract microbiotas as the source of the lung and gastric microbiotas in healthy individuals. MBio. 2015; 6:e00037. PMID: 25736890.20. Salter SJ, Cox MJ, Turek EM, Calus ST, Cookson WO, Moffatt MF, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014; 12:87. PMID: 25387460.21. Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. 16S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome. 2015; 3:26. PMID: 26120470.22. Davis IJ, Bull C, Horsfall A, Morley I, Harris S. The Unculturables: targeted isolation of bacterial species associated with canine periodontal health or disease from dental plaque. BMC Microbiol. 2014; 14:196. PMID: 25085267.23. Ohno M, Okano I, Watsuji T, Kakinuma T, Ueda K, Beppu T. Establishing the independent culture of a strictly symbiotic bacterium Symbiobacterium thermophilum from its supporting Bacillus strain. Biosci Biotechnol Biochem. 1999; 63:1083–1090. PMID: 10427695.24. Hiergeist A, Glasner J, Reischl U, Gessner A. Analyses of Intestinal Microbiota: Culture versus Sequencing. ILAR J. 2015; 56:228–240. PMID: 26323632.25. Maukonen J, Simoes C, Saarela M. The currently used commercial DNA-extraction methods give different results of clostridial and actinobacterial populations derived from human fecal samples. FEMS Microbiol Ecol. 2012; 79:697–708. PMID: 22098067.26. Willner D, Daly J, Whiley D, Grimwood K, Wainwright CE, Hugenholtz P. Comparison of DNA extraction methods for microbial community profiling with an application to pediatric bronchoalveolar lavage samples. PLoS One. 2012; 7:e34605. PMID: 22514642.27. Kennedy NA, Walker AW, Berry SH, Duncan SH, Farquarson FM, Louis P, et al. The impact of different DNA extraction kits and laboratories upon the assessment of human gut microbiota composition by 16S rRNA gene sequencing. PLoS One. 2014; 9:e88982. PMID: 24586470.28. Beck JM. ABCs of the lung microbiome. Ann Am Thorac Soc. 2014; 11(Suppl 1):S3–S6. PMID: 24437402.29. Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, et al. Diversity of the human intestinal microbial flora. Science. 2005; 308:1635–1638. PMID: 15831718.30. Zoetendal EG, Vaughan EE, de Vos WM. A microbial world within us. Mol Microbiol. 2006; 59:1639–1650. PMID: 16553872.31. Fournier PE, Lagier JC, Dubourg G, Raoult D. From culturomics to taxonomogenomics: a need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe. 2015; 36:73–78. PMID: 26514403.32. Lagier JC, Hugon P, Khelaifia S, Fournier PE, La Scola B, Raoult D. The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin Microbiol Rev. 2015; 28:237–264. PMID: 25567229.33. Lagier JC, Million M, Hugon P, Armougom F, Raoult D. Human gut microbiota: repertoire and variations. Front Cell Infect Microbiol. 2012; 2:136. PMID: 23130351.34. Samb-Ba B, Mazenot C, Gassama-Sow A, Dubourg G, Richet H, Hugon P, et al. MALDI-TOF Identification of the Human Gut Microbiome in People with and without Diarrhea in Senegal. Plos One. 2014; 9:e87419. PMID: 24784934.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Taxonomic Identification of Bacillus Species Using Matrix-Assisted Laser Desorption/Ionization-Time of Flight Mass Spectrometry
- Comparison of MALDI-TOF MS, Housekeeping Gene Sequencing, and 16S rRNA Gene Sequencing for Identification of Aeromonas Clinical Isolates
- The Diagnostic Value of Bronchoalveolar lavage fluid microscopic study and PCR in Pulmonary tuberculosis
- Changes of the cellularities in the bronchoalveolar lavage fluid of the experimental silicosis
- Evaluation of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry for Identification of Aerobic Bacteria in a Clinical Microbiology Laboratory