J Korean Acad Prosthodont.
2017 Oct;55(4):361-371. 10.4047/jkap.2017.55.4.361.
Regulation of human gingival fibroblast gene expression on microgrooves: A DNA microarray study
- Affiliations
-
- 1Department of Dentistry, Graduate School, Kyung Hee University, Seoul, Republic of Korea.
- 2Department of Biomaterials & Prosthodontics, Kyung Hee University Hospital at Gangdong, School of Dentistry, Kyung Hee University, Seoul, Republic of Korea. ysprosth@hanmail.net
- KMID: 2393907
- DOI: http://doi.org/10.4047/jkap.2017.55.4.361
Abstract
- PURPOSE
We aimed to investigate the gene expression of human gingival fibroblasts on microgroove surface using DNA microarray.
MATERIALS AND METHODS
Microgrooves were applied on grade II titanium discs to have 0/0 µm (NE0, control group), 60/10 µm (E60/10, experimental group) of respective width/depth by photolithography. The entire surface of the microgrooved Ti substrata was further acid etched and used as the two experimental groups in this study. Human gingival fibroblasts were cultured in the experimental group and the control group, and total RNA was extracted. The oligonucleotide microarray was performed to confirm the changes of various gene expression levels between experimental group and control group. Changes of gene expression level were determined at the pathway level by mapping the expression results of DNA chips, using the KEGG (Kyoto Encyclopedia of Genes and Genomes) pathway analysis.
RESULTS
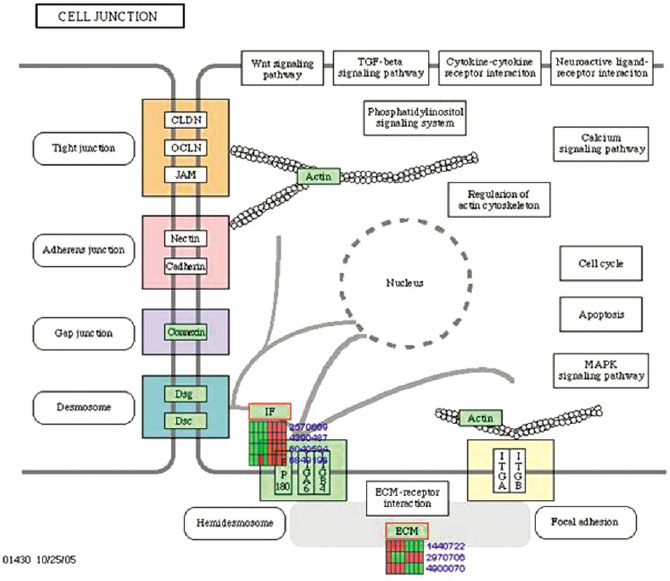
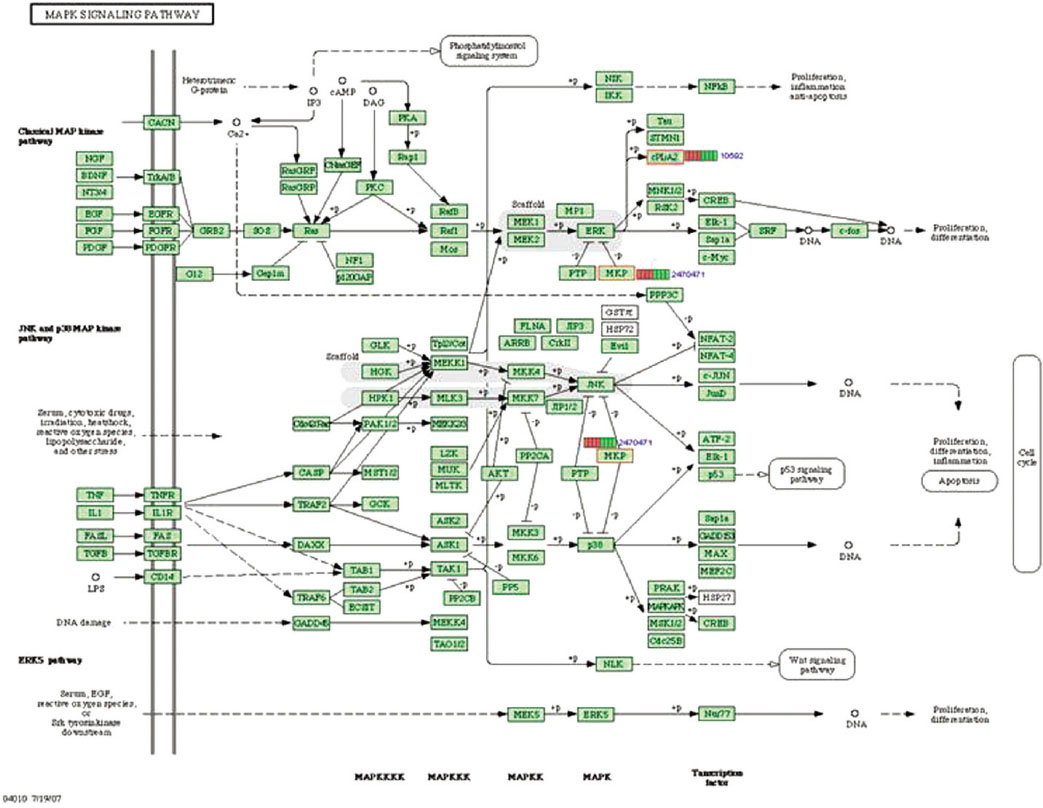
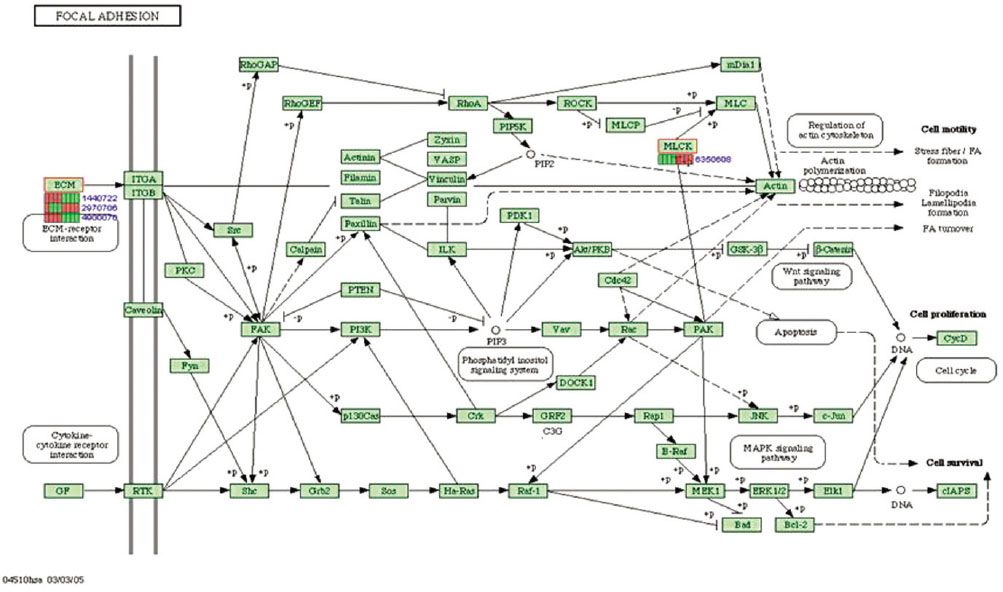
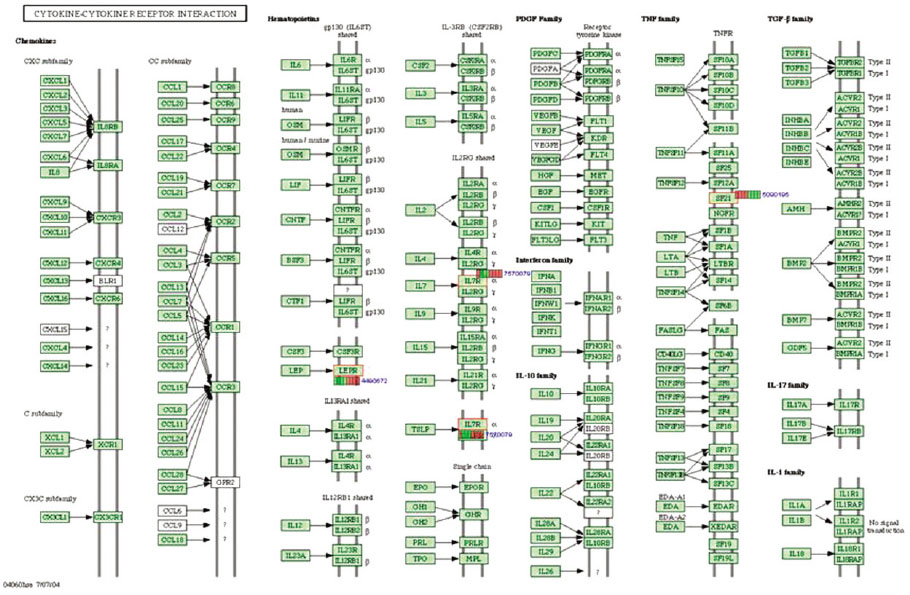
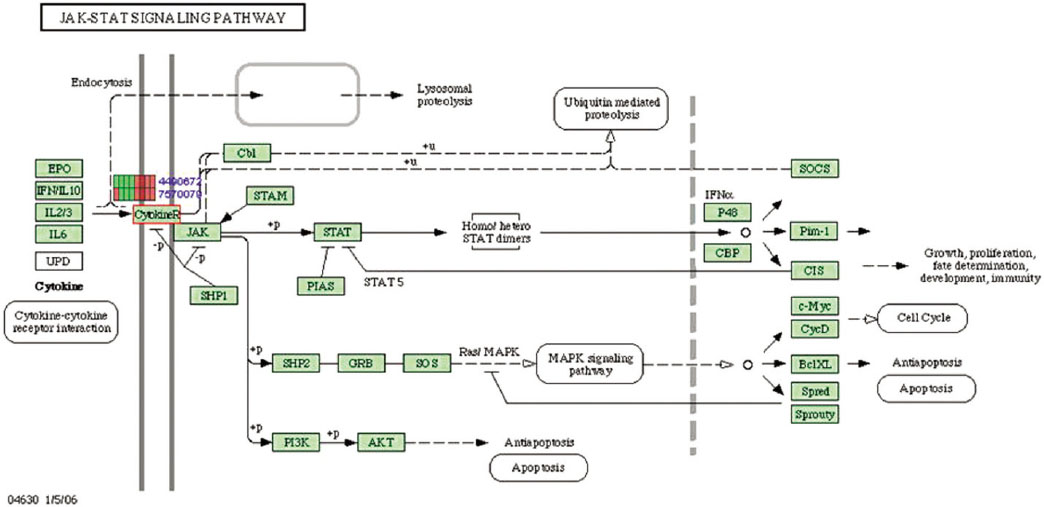
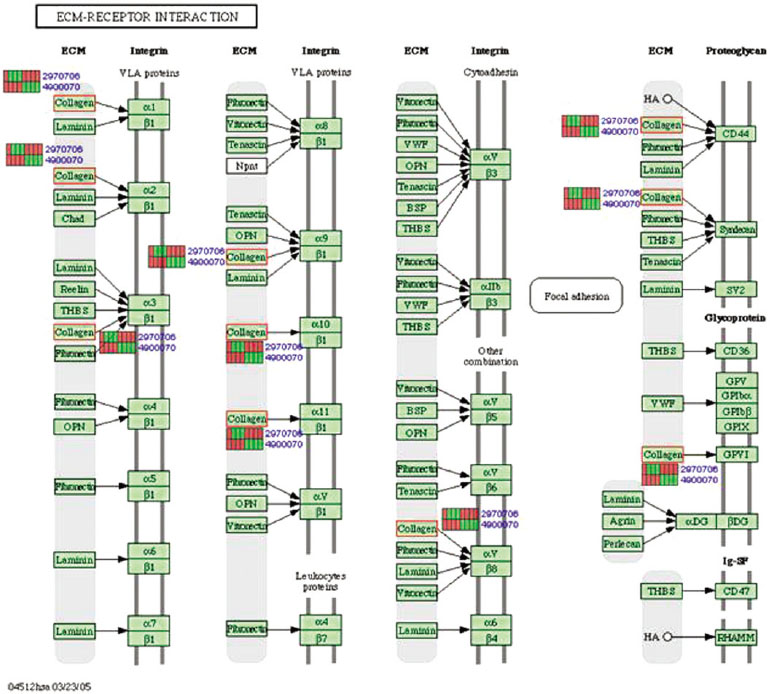
Gene expression levels on E60/10 and NE0 were analyzed, there were 123 genes showing significant differences in expression more than 1.5 times on E60/10 microgrooved surface compared to NE0 surface, and 19 genes showing significant differences in expression more than 2 times. The KEGG pathway analysis confirmed the changes in gene expression levels under experimental conditions. Cell signaling, proliferation, and activity among the various gene expression results were identified.
CONCLUSION
Microgrooved surfaces induce gene expression changes and related cell signaling. According to the results of this study, microgrooves can be used as the surface of various biomaterials which need to improve cell activity through gene expression changes and activation of cell signaling.
MeSH Terms
Figure
Reference
-
1. Kasemo B, Lausmaa J. Surface science aspects on inorganic biomaterials. CRC critical reviews in biocompatibility. Boca Raton FL: CRC Press;1986. p. 335–380.2. Albrektsson T, Wennerberg A. Oral implant surfaces: Part 1-review focusing on topographic and chemical properties of different surfaces and in vivo responses to them. Int J Prosthodont. 2004; 17:536–543.3. Morra M, Cassinelli C, Bruzzone G, Carpi A, Di Santi G, Giardino R, Fini M. Surface chemistry effects of topographic modification of titanium dental implant surfaces: 1. Surface analysis. Int J Oral Maxillofac Implants. 2003; 18:40–45.4. Zhao L, Mei S, Chu PK, Zhang Y, Wu Z. The influence of hierarchical hybrid micro/nano-textured titanium surface with titania nanotubes on osteoblast functions. Biomaterials. 2010; 31:5072–5082.
Article5. Lee SW, Kim SY, Rhyu IC, Chung WY, Leesungbok R, Lee KW. Influence of microgroove dimension on cell behavior of human gingival fibroblasts cultured on titanium substrata. Clin Oral Implants Res. 2009; 20:56–66.
Article6. Chou L, Firth JD, Uitto VJ, Brunette DM. Substratum surface topography alters cell shape and regulates fibronectin mRNA level, mRNA stability, secretion and assembly in human fibroblasts. J Cell Sci. 1995; 108:1563–1573.
Article7. Jockusch BM, Bubeck P, Giehl K, Kroemker M, Moschner J, Rothkegel M, Rüdiger M, Schlüter K, Stanke G, Winkler J. The molecular architecture of focal adhesions. Annu Rev Cell Dev Biol. 1995; 11:379–416.
Article8. Pankov R, Yamada KM. Fibronectin at a glance. J Cell Sci. 2002; 115:3861–3863.
Article9. Park SJ, Leesungbok R, Ahn SJ, Im BJ, Lee DY, Jee YJ, Yoon JH, Cui T, Lee SC, Lee SW. Effect of microgrooves and fibronectin conjugation on the osteoblast marker gene expression and differentiation. J Adv Prosthodont. 2015; 7:496–505.
Article10. Lai EC. Notch signaling: control of cell communication and cell fate. Development. 2004; 131:965–973.
Article11. Halper J, Kjaer M. Basic components of connective tissues and extracellular matrix: elastin, fibrillin, fibulins, fibrinogen, fibronectin, laminin, tenascins and thrombospondins. Progress in Heritable Soft Connective Tissue Diseases. Springer;2014. p. 31–47.12. Yun-Feng W, Matsuo N, Sumiyoshi H, Yoshioka H. Sp7/Osterix up-regulates the mouse pro-alpha3(V) collagen gene (Col5a3) during the osteoblast differentiation. Biochem Biophys Res Commun. 2010; 394:503–508.
Article13. Kadler KE, Hill A, Canty-Laird EG. Collagen fibrillogenesis: fibronectin, integrins, and minor collagens as organizers and nucleators. Curr Opin Cell Biol. 2008; 20:495–501.
Article14. Mitra SK, Hanson DA, Schlaepfer DD. Focal adhesion kinase: in command and control of cell motility. Nat Rev Mol Cell Biol. 2005; 6:56–68.
Article15. Zhang W, Liu HT. MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 2002; 12:9–18.
Article16. Yang P, Roy SK. A novel mechanism of FSH regulation of DNA synthesis in the granulosa cells of hamster preantral follicles: involvement of a protein kinase C-mediated MAP kinase 3/1 self-activation loop. Biol Reprod. 2006; 75:149–157.
Article17. Balko JM, Schwarz LJ, Bhola NE, Kurupi R, Owens P, Miller TW, Go′mez H, Cook RS, Arteaga CL. Activation of MAPK pathways due to DUSP4 loss promotes cancer stem cell-like phenotypes in basal-like breast cancer. Cancer Res. 2013; 73:6346–6358.
Article18. Leonard WJ, Lin JX. Cytokine receptor signaling pathways. J Allergy Clin Immunol. 2000; 105:877–888.
Article19. Baumann H, Morella KK, White DW, Dembski M, Bailon PS, Kim H, Lai CF, Tartaglia LA. The full-length leptin receptor has signaling capabilities of interleukin 6-type cytokine receptors. Proc Natl Acad Sci USA. 1996; 93:8374–8378.
Article20. Peschon JJ, Morrissey PJ, Grabstein KH, Ramsdell FJ, Maraskovsky E, Gliniak BC, Park LS, Ziegler SF, Williams DE, Ware CB, Meyer JD, Davison BL. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994; 180:1955–1960.
Article21. Mi S, Lee X, Hu Y, Ji B, Shao Z, Yang W, Huang G, Walus L, Rhodes K, Gong BJ, Miller RH, Pepinsky RB. Death receptor 6 negatively regulates oligodendrocyte survival, maturation and myelination. Nat Med. 2011; 17:816–821.
Article22. Versteeg HH, Spek CA, Peppelenbosch MP, Richel DJ. Tissue factor and cancer metastasis: the role of intracellular and extracellular signaling pathways. Mol Med. 2004; 10:6–11.
Article23. Rawlings JS, Rosler KM, Harrison DA. The JAK/STAT signaling pathway. J Cell Sci. 2004; 117:1281–1283.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Surface microgrooves of thirty micrometers in width on titanium substrata enhance proliferation and alter gene expression of cultured human gingival fibroblasts
- Profiling of genes in healthy hGF, aging hGF, healthy hPDLF and inflammatory hPDLF by DNA microarray
- Gene expression after the application of the fluid-induced shear stress on the gingival fibroblast
- Micropatterned grooves and acid-etching on titanium substrata alter viability and gene expression of adhered human gingival fibroblasts: A pilot study
- A Comparative Study of Gene Expression Patterns of Periodontal Ligament Cells and Gingival Fibroblasts using the cDNA Microarray