Tuberc Respir Dis.
2008 Dec;65(6):495-503.
Identification of DNA Methylation Markers for NSCLC Using Hpall-Mspl Methylation Microarray
- Affiliations
-
- 1Department of Internal Medicine, Konyang University School of Medicine, Daejeon, Korea. sk1609@hanmail.net
- 2Department of Chest Surgery, Konyang University School of Medicine, Daejeon, Korea.
- 3Department of Pathology, Konyang University School of Medicine, Daejeon, Korea.
- 4Department of Radiology, Konyang University School of Medicine, Daejeon, Korea.
Abstract
-
BACKGROUND: Epigenetic alterations in certain genes are now known as at least important as genetic mutation in pathogenesis of cancer. Especially abnormal hypermethylation in or near promoter region of tumor suppressor genes (TSGs) are known to result in gene silencing and loss of gene function eventually. The authors tried to search for new lung cancer-specific TSGs which have CpG islands and HpaII sites, and are thought to be involved in carcinogenesis by epigenetic mechanism.
METHODS
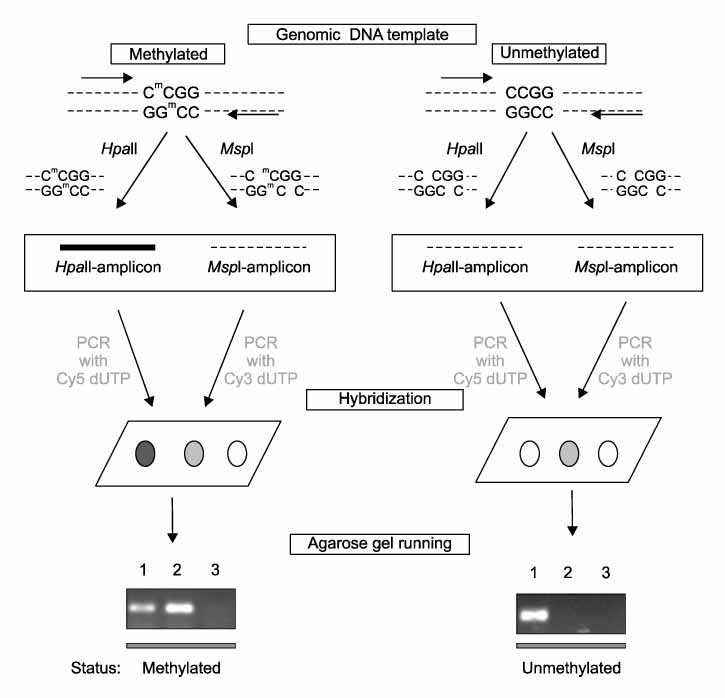
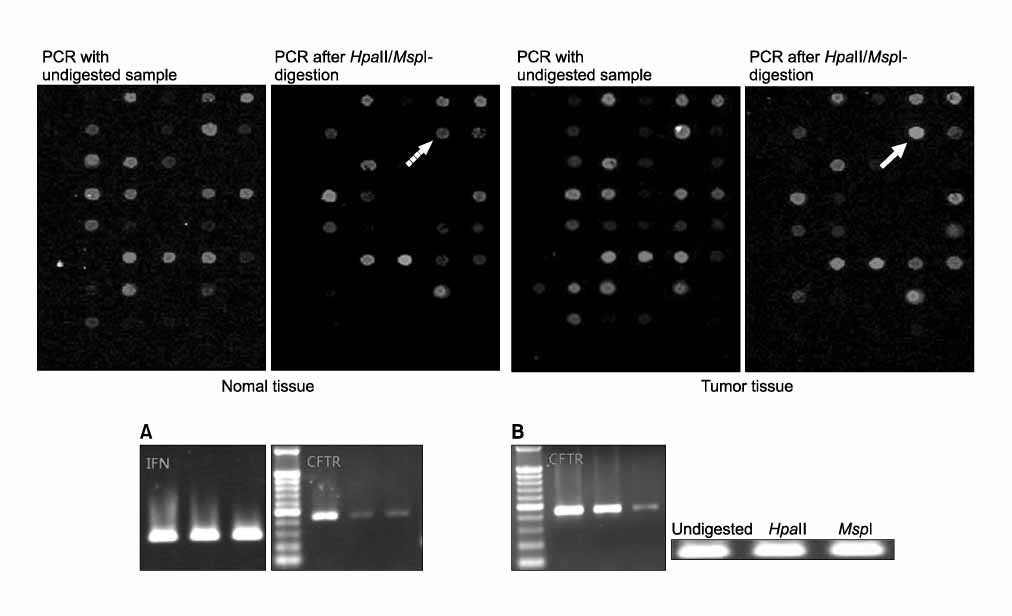
Tumor tissue and corresponding adjacent normal tissue were obtained from 10 patients who diagnosed with non small cell lung cancer (NSCLC) and underwent surgery in Konyang university hospital in 2005. Methylation profiles of promoter region of 21 genes in tumor tissue & non-tumor tissue were examined with HpaII-MspI methylation microarray (Methyl-Scan DNA chip(R), Genomic tree, Inc, South Korea). The rates of hypermethylation were compared in tumor and non-tumor group, and as a normal control, we obtained lung tissue from two young patients with pneumothorax during bullectomies, methylation profiles were examined in the same way.
RESULTS
Among the 21 genes, 10 genes were commonly methylated in tumor, non-tumor, and control group. The 6 genes of APC, AR, RAR-b, HTR1B, EPHA3, and CFTR, among the rest of 11 genes were not methylated in control, and more frequently hypermethylated in tumor tissue than non-tumor tissue.
CONCLUSION
In the present study, HTR1B, EPHA3, and CFTR are suggested as possible novel TSGs of NSCLC by epigenetic mechanism.
MeSH Terms
Figure
Reference
-
1. Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007. 57:43–66.2. Ellis JR, Gleeson FV. Lung cancer screening. Br J Radiol. 2001. 74:478–485.3. Marcus PM. Lung cancer screening: an update. J Clin Oncol. 2001. 19:83S–86S.4. Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002. 3:415–428.5. Worm J, Guldberg P. DNA methylation: an epigenetic pathway to cancer and a promising target for anticancer therapy. J Oral Pathol Med. 2002. 31:443–449.6. Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol. 2005. 2:Suppl 1. S4–S11.7. Esteller M, Corn PG, Baylin SB, Herman JG. A gene hypermethylation profile of human cancer. Cancer Res. 2001. 61:3225–3229.8. Zöchbauer-Müller S, Minna JD, Gazdar AF. Aberrant DNA methylation in lung cancer: biological and clinical implications. Oncologist. 2002. 7:451–457.9. Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, et al. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000. 60:5954–5958.10. Park JC, Chae YK, Son CH, Kim MS, Lee J, Ostrow K, et al. Epigenetic silencing of human T (brachyury homologue) gene in non-small-cell lung cancer. Biochem Biophys Res Commun. 2008. 365:221–226.11. Gu J, Berman D, Lu C, Wistuba II, Roth JA, Frazier M, et al. Aberrant promoter methylation profile and association with survival in patients with non-small cell lung cancer. Clin Cancer Res. 2006. 12:7329–7338.12. Harden SV, Tokumaru Y, Westra WH, Goodman S, Ahrendt SA, Yang SC, et al. Gene promoter hypermethylation in tumors and lymph nodes of stage I lung cancer patients. Clin Cancer Res. 2003. 9:1370–1375.13. Kim DH, Kim JS, Ji YI, Shim YM, Kim H, Han J, et al. Hypermethylation of RASSF1A promoter is associated with the age at starting smoking and a poor prognosis in primary non-small cell lung cancer. Cancer Res. 2003. 63:3743–3746.14. Adorján P, Distler J, Lipscher E, Model F, Müller J, Pelet C, et al. Tumour class prediction and discovery by microarray-based DNA methylation analysis. Nucleic Acids Res. 2002. 30:e21.15. Shi H, Maier S, Nimmrich I, Yan PS, Caldwell CW, Olek A, et al. Oligonucleotide-based microarray for DNA methylation analysis: principles and applications. J Cell Biochem. 2003. 88:138–143.16. Yan PS, Chen CM, Shi H, Rahmatpanah F, Wei SH, Caldwell CW, et al. Dissecting complex epigenetic alterations in breast cancer using CpG island microarrays. Cancer Res. 2001. 61:8375–8380.17. Fukasawa M, Kimura M, Morita S, Matsubara K, Yamanaka S, Endo C, et al. Microarray analysis of promoter methylation in lung cancers. J Hum Genet. 2006. 51:368–374.18. Hatada I, Kato A, Morita S, Obata Y, Nagaoka K, Sakurada A, et al. A microarray-based method for detecting methylated loci. J Hum Genet. 2002. 47:448–451.19. Hou P, Ji M, He N, Lu Z. Microarray-based method to evaluate the accuracy of restriction endonucleases HpaII and MspI. Biochem Biophys Res Commun. 2004. 314:110–117.20. Takai D, Yagi Y, Wakazono K, Ohishi N, Morita Y, Sugimura T, et al. Silencing of HTR1B and reduced expression of EDN1 in human lung cancers, revealed by methylation-sensitive representational difference analysis. Oncogene. 2001. 20:7505–7513.21. Kim JS, Han J, Shim YM, Park J, Kim DH. Aberrant methylation of H-cadherin (CDH13) promoter is associated with tumor progression in primary nonsmall cell lung carcinoma. Cancer. 2005. 104:1825–1833.22. Sato M, Mori Y, Sakurada A, Fujimura S, Horii A. The H-cadherin (CDH13) gene is inactivated in human lung cancer. Hum Genet. 1998. 103:96–101.23. Toyooka KO, Toyooka S, Virmani AK, Sathyanarayana UG, Euhus DM, Gilcrease M, et al. Loss of expression and aberrant methylation of the CDH13 (H-cadherin) gene in breast and lung carcinomas. Cancer Res. 2001. 61:4556–4560.24. Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, Park JY. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer. 2007. 110:2785–2792.25. Bunn PA Jr, Helfrich BA, Brenner DG, Chan DC, Dykes DJ, Cohen AJ, et al. Effects of recombinant neutral endopeptidase (EC 3.4.24.11) on the growth of lung cancer cell lines in vitro and in vivo. Clin Cancer Res. 1998. 4:2849–2858.26. Cohen AJ, Franklin WA, Magill C, Sorenson J, Miller YE. Low neutral endopeptidase levels in bronchoalveolar lavage fluid of lung cancer patients. Am J Respir Crit Care Med. 1999. 159:907–910.27. Kristiansen G, Schluns K, Yongwei Y, Dietel M, Petersen I. CD10 expression in non-small cell lung cancer. Anal Cell Pathol. 2002. 24:41–46.28. Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001. 293:1068–1070.29. Sachan M, Raman R. Developmental methylation of the regulatory region of HoxB5 gene in mouse correlates with its tissue-specific expression. Gene. 2006. 380:151–158.30. Yung RL, Julius A. Epigenetics, aging, and autoimmunity. Autoimmunity. 2008. 41:329–335.31. Tsoli E, Gorgoulis VG, Zacharatos P, Kotsinas A, Mariatos G, Kastrinakis NG, et al. Low levels of p27 in association with deregulated p53-pRb protein status enhance tumor proliferation and chromosomal instability in non-small cell lung carcinomas. Mol Med. 2001. 7:418–429.32. Virmani AK, Tsou JA, Siegmund KD, Shen LY, Long TI, Laird PW, et al. Hierarchical clustering of lung cancer cell lines using DNA methylation markers. Cancer Epidemiol Biomarkers Prev. 2002. 11:291–297.33. Winters ZE. P53 pathways involving G2 checkpoint regulators and the role of their subcellular localisation. J R Coll Surg Edinb. 2002. 47:591–598.34. Zöchbauer-Müller S, Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001. 61:249–255.35. Tsou JA, Hagen JA, Carpenter CL, Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002. 21:5450–5461.36. Jacquot J, Tabary O, Le Rouzic P, Clement A. Airway epithelial cell inflammatory signalling in cystic fibrosis. Int J Biochem Cell Biol. 2008. 40:1703–1715.37. Cantin AM, Bilodeau G, Ouellet C, Liao J, Hanrahan JW. Oxidant stress suppresses CFTR expression. Am J Physiol Cell Physiol. 2006. 290:C262–C270.38. Shaulian E, Karin M. AP-1 in cell proliferation and survival. Oncogene. 2001. 20:2390–2400.39. Karamouzis MV, Konstantinopoulos PA, Papavassiliou AG. The activator protein-1 transcription factor in respiratory epithelium carcinogenesis. Mol Cancer Res. 2007. 5:109–120.40. Brantley DM, Cheng N, Thompson EJ, Lin Q, Brekken RA, Thorpe PE, et al. Soluble Eph A receptors inhibit tumor angiogenesis and progression in vivo. Oncogene. 2002. 21:7011–7026.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- DNA Methylation in Development
- DNA Methylation in Lung Cancer
- A Visualization Tool for Computational Analysis of DNA Methylation Level Using Bisulfite Sequencing Data
- DNA methylation: a cause and consequence of type 2 diabetes
- DNA Methylation in Brain and Liver Tissues of Mice Infected with Scrapie Agent