Anat Cell Biol.
2010 Mar;43(1):1-14. 10.5115/acb.2010.43.1.1.
Genomics and proteomics in stem cell research: the road ahead
- Affiliations
-
- 1LCDI-BRC Joint Genome Center, Lee Gil Ya Cancer and Diabetes Institute, Gachon University of Medicine and Science, Incheon, Korea.
- 2Joint Proteomics Laboratory, Ludwig Institute for Cancer Research & The Walter and Eliza Hall Institute of Medical Research, University of Melbourne, Melbourne, Australia.
- 3Center for Genomics and Proteomics & Stem Cell Core Facility, Lee Gil Ya Cancer and Diabetes Institute, Gachon University of Medicine and Science, Incheon, Korea. bhlee@gachon.ac.kr
- KMID: 2168892
- DOI: http://doi.org/10.5115/acb.2010.43.1.1
Abstract
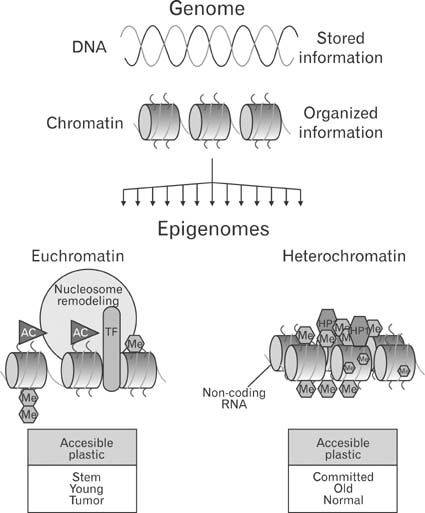
- Stem cell research has been widely studied over the last few years and has attracted increasing attention from researchers in all fields of medicine due to its potential to treat many previously incurable diseases by replacing damaged cells or tissues. As illustrated by hematopoietic stem research, understanding stem cell differentiation at molecular levels is essential for both basic research and for clinical applications of stem cells. Although multiple integrative analyses, such as genomics, epigenomics, transcriptomics and proteomics, are required to understand stem cell biology, proteomics has a unique position in stem cell research. For example, several major breakthroughs in HSC research were due to the identification of proteins such as colony-stimulating factors (CSFs) and cell-surface CD molecules. In 2007, the Human Proteome Organization (HUPO) and the International Society for Stem Cell Research (ISSCR) launched the joint Proteome Biology of Stem Cells Initiative. A systematic proteomics approach to understanding stem cell differentiation will shed new light on stem cell biology and accelerate clinical applications of stem cells.
Keyword
MeSH Terms
Figure
Reference
-
1. Agaton C, Galli J, Höidén Guthenberg I, et al. Affinity proteomics for systematic protein profiling of chromosome 21 gene products in human tissues. Mol Cell Proteomics. 2003. 2:405–414.2. Ahmad K, Henikoff S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Molecular cell. 2002. 9:1191–1200.3. Alberts B. Molecular biology of the cell. 2002. 4th edn. New York: Garland Science.4. Arney KL, Fisher AG. Epigenetic aspects of differentiation. J Cell Sci. 2004. 117:4355–4363.5. Arrell DK, Niederlander NJ, Faustino RS, Behfar A, Terzic A. Cardioinductive network guiding stem cell differentiation revealed by proteomic cartography of tumor necrosis factor alpha-primed endodermal secretome. Stem Cells. 2008. 26:387–400.6. Avilion AA, Nicolis SK, Pevny LH, Perez L, Vivian N, Lovell-Badge R. Multipotent cell lineages in early mouse development depend on SOX2 function. Genes Dev. 2003. 17:126–140.7. Baba M, Hasegawa H, Nakayabu M, et al. Establishment and characteristics of a gastric cancer cell line (HuGC-OOHIRA) producing high levels of G-CSF, GM-CSF, and IL-6: the presence of autocrine growth control by G-CSF. Am J Hematol. 1995. 49:207–215.8. Bannister AJ, Zegerman P, Partridge JF, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001. 410:120–124.9. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004. 116:281–297.10. Bartel DP, Chen CZ. Micromanagers of gene expression: the potentially widespread influence of metazoan microRNAs. Nat Rev Genet. 2004. 5:396–400.11. Baudino TA, Cleveland JL. The Max network gone mad. Mol Cell Biol. 2001. 21:691–702.12. Bentley GA, Boulot G, Chitarra V. Cross-reactivity in antibody-antigen interactions. Res Immunol. 1994. 145:45–48.13. Bernstein BE, Mikkelsen TS, Xie X, et al. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell. 2006. 125:315–326.14. Bhutani N, Brady JJ, Damian M, Sacco A, Corbel SY, Blau HM. Reprogramming towards pluripotency requires AID-dependent DNA demethylation. Nature. 2010. 463:1042–1047.15. Bird A. DNA methylation patterns and epigenetic memory. Genes Development. 2002. 16:6–21.16. Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003. 72:291–336.17. Blanchette M, Bataille AR, Chen X, et al. Genome-wide computational prediction of transcriptional regulatory modules reveals new insights into human gene expression. Genome Res. 2006. 16:656–668.18. Boyer LA, Lee TI, Cole MF, et al. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005. 122:947–956.19. Bradley TR, Metcalf D. The growth of mouse bone marrow cells in vitro. Aust J Exp Biol Med Sci. 1966. 44:287–299.20. Burns CE, Zon LI. Portrait of a stem cell. Dev Cell. 2002. 3:612–613.21. Byrne JA, Pedersen DA, Clepper LL, et al. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007. 450:497–502.22. Cai J, Weiss ML, Rao MS. In search of "stemness". Exp Hematol. 2004. 32:585–598.23. Chambers I, Silva J, Colby D, et al. Nanog safeguards pluripotency and mediates germline development. Nature. 2007. 450:1230–1234.24. Chang HY, Thomson JA, Chen X. Microarray analysis of stem cells and differentiation. Methods Enzymol. 2006. 420:225–254.25. Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004. 303:83–86.26. Cheng LC, Tavazoie M, Doetsch F. Stem cells: from epigenetics to microRNAs. Neuron. 2005. 46:363–367.27. Clarke MF, Fuller M. Stem cells and cancer: two faces of eve. Cell. 2006. 124:1111–1115.28. Cui Q, Yu Z, Purisima EO, Wang E. Principles of microRNA regulation of a human cellular signaling network. Mol Syst Biol. 2006. 2:46.29. Davidson EH. The regulatory genome: gene regulatory networks in development and evolution. 2006. New edn. Oxford Boston: Elsevier / Academic Press.30. Dou Y, Gorovsky MA. Phosphorylation of linker histone H1 regulates gene expression in vivo by creating a charge patch. Molecular Cell. 2000. 6:225–231.31. Dover J, Schneider J, Tawiah-Boateng MA, et al. Methylation of histone H3 by COMPASS requires ubiquitination of histone H2B by Rad6. J Biol Chem. 2002. 277:28368–28371.32. Draper JS, Smith K, Gokhale P, et al. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2004. 22:53–54.33. Edman P. Phenylthiohydantoins in protein analysis. Ann N Y Acad Sci. 1960. 88:602–610.34. Elliott RL, Blobe GC. Role of transforming growth factor Beta in human cancer. J Clin Oncol. 2005. 23:2078–2093.35. Elliott ST, Crider DG, Garnham CP, Boheler KR, Van Eyk JE. Two-dimensional gel electrophoresis database of murine R1 embryonic stem cells. Proteomics. 2004. 4:3813–3832.36. Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006. 7:21–33.37. Feldman N, Gerson A, Fang J, et al. G9a-mediated irreversible epigenetic inactivation of Oct-3/4 during early embryogenesis. Nat Cell Biol. 2006. 8:188–194.38. Fischle W, Wang Y, Allis CD. Binary switches and modification cassettes in histone biology and beyond. Nature. 2003. 425:475–479.39. Fortunel NO, Otu HH, Ng HH, et al. Comment on " 'Stemness': transcriptional profiling of embryonic and adult stem cells" and "a stem cell molecular signature". Science. 2003. 302:393.40. Gandhi TK, Zhong J, Mathivanan S, et al. Analysis of the human protein interactome and comparison with yeast, worm and fly interaction datasets. Nat Genet. 2006. 38:285–293.41. Garcia BA, Pesavento JJ, Mizzen CA, Kelleher NL. Pervasive combinatorial modification of histone H3 in human cells. Nat Methods. 2007. 4:487–489.42. Gasson JC, Weisbart RH, Kaufman SE, et al. Purified human granulocyte-macrophage colony-stimulating factor: direct action on neutrophils. Science. 1984. 226:1339–1342.43. He L, He X, Lim LP, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007. 447:1130–1134.44. Heck AJ, Mummery C, Whetton AD, et al. Proteome biology of stem cells. Stem Cell Res. 2007. 1:7–8.45. Hopkins AL, Groom CR. The druggable genome. Nat Rev Drug Discov. 2002. 1:727–730.46. Howard ML, Davidson EH. cis-Regulatory control circuits in development. Dev Biol. 2004. 271:109–118.47. Hunt DF, Yates JR 3rd, Shabanowitz J, Winston S, Hauer CR. Protein sequencing by tandem mass spectrometry. Proc Natl Acad Sci U S A. 1986. 83:6233–6237.48. Ivanova NB, Dimos JT, Schaniel C, Hackney JA, Moore KA, Lemischka IR. A stem cell molecular signature. Science. 2002. 298:601–604.49. Jacobs JM, Waters KM, Kathmann LE, et al. The mammary epithelial cell secretome and its regulation by signal transduction pathways. J Proteome Res. 2008. 7:558–569.50. Jenuwein T, Allis CD. Translating the histone code. Science. 2001. 293:1074–1080.51. Johnston RJ Jr, Chang S, Etchberger JF, Ortiz CO, Hobert O. MicroRNAs acting in a double-negative feedback loop to control a neuronal cell fate decision. Proc Natl Acad Sci U S A. 2005. 102:12449–12454.52. Jones PA, Takai D. The role of DNA methylation in mammalian epigenetics. Science. 2001. 293:1068–1070.53. Judson RL, Babiarz JE, Venere M, Blelloch R. Embryonic stem cell-specific microRNAs promote induced pluripotency. Nat Biotechnol. 2009. 27:459–461.54. Kang DJ, Oh SO, Ahn SM, Lee BH, Moon MH. Proteomic analysis of exosomes from human neural stem cells by flow field-flow fractionation and nanoflow liquid chromatography-tandem mass spectrometry. J Proteome Res. 2008. 7:3475–3480.55. Kang YK, Koo DB, Park JS, et al. Aberrant methylation of donor genome in cloned bovine embryos. Nat Genet. 2001. 28:173–177.56. Khwaja FW, Svoboda P, Reed M, Pohl J, Pyrzynska B, Van Meir EG. Proteomic identification of the wt-p53-regulated tumor cell secretome. Oncogene. 2006. 25:7650–7661.57. Kim H, Hahn M, Grabowski P, et al. The Bacillus subtilis spore coat protein interaction network. Mol Microbiol. 2006. 59:487–502.58. Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron. 2003. 38:17–31.59. Klose RJ, Sarraf SA, Schmiedeberg L, McDermott SM, Stancheva I, Bird AP. DNA binding selectivity of MeCP2 due to a requirement for A/T sequences adjacent to methyl-CpG. Mol Cell. 2005. 19:667–678.60. Kratchmarova I, Blagoev B, Haack-Sorensen M, Kassem M, Mann M. Mechanism of divergent growth factor effects in mesenchymal stem cell differentiation. Science. 2005. 308:1472–1477.61. Krijgsveld J, Whetton AD, Lee BH, et al. Proteome biology of stem cells: a new joint HUPO and ISSCR initiative. Mol Cell Proteomics. 2008. 7:204–205.62. Krishna RG, Wold F. Post-translational modification of proteins. Adv Enzymol Relat Areas Mol Biol. 1993. 67:265–298.63. Lee JH, Hart SRL, Skalnik DG. Histone deacetylase activity is required for embryonic stem cell differentiation. Genesis. 2004. 38:32–38.64. Levchenko A. Proteomics takes stem cell analyses to another level. Nat Biotechnol. 2005. 23:828–830.65. Lim LP, Lau NC, Garrett-Engele P, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005. 433:769–773.66. Liu C, Zhao X. MicroRNAs in adult and embryonic neurogenesis. Neuromolecular Med. 2009. 11:141–152.67. Luger K, Mäder AW, Richmond RK, Sargent DF, Richmond TJ. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature. 1997. 389:251–260.68. Luzi E, Marini F, Carbonell SS, Tognarini I, Galli G, Brandi ML. Osteogenic differentiation of human adipose tissue-derived stem cells is modulated by the miR-26a targeting the SMAD1 transcription factor. J Bone Miner Res. 2008. 23:287–295.69. Maitra A, Arking DE, Shivapurkar N, et al. Genomic alterations in cultured human embryonic stem cells. Nat Genet. 2005. 37:1099–1103.70. Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005. 15:163–176.71. Marx J. Cancer research. Mutant stem cells may seed cancer. Science. 2003. 301:1308–1310.72. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. 2006. 15(Spec No 1):R17–R29.73. Metcalf D. The Florey Lecture, 1991. The colony-stimulating factors: discovery to clinical use. Philos Trans R Soc Lond B Biol Sci. 1991. 333:147–173.74. Nichols J, Zevnik B, Anastassiadis K, et al. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 1998. 95:379–391.75. Nightingale KP, O'Neill LP, Turner BM. Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev. 2006. 16:125–136.76. Nomura H, Imazeki I, Oheda M, et al. Purification and characterization of human granulocyte colony-stimulating factor (G-CSF). EMBO J. 1986. 5:871–876.77. Ong SE, Mittler G, Mann M. Identifying and quantifying in vivo methylation sites by heavy methyl SILAC. Nat Methods. 2004. 1:119–126.78. O'Shea JJ, Gadina M, Schreiber RD. Cytokine signaling in 2002: new surprises in the Jak/Stat pathway. Cell. 2002. 109:Suppl. S121–S131.79. Pardal R, Clarke MF, Morrison SJ. Applying the principles of stem-cell biology to cancer. Nat Rev Cancer. 2003. 3:895–902.80. Pasquinelli AE, Hunter S, Bracht J. MicroRNAs: a developing story. Curr Opin Genet Dev. 2005. 15:200–205.81. Pesavento JJ, Kim YB, Taylor GK, Kelleher NL. Shotgun annotation of histone modifications: a new approach for streamlined characterization of proteins by top down mass spectrometry. J Am Chem Soc. 2004. 126:3386–3387.82. Plasterk RH. Micro RNAs in animal development. Cell. 2006. 124:877–881.83. Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004. 432:226–230.84. Ramalho-Santos M, Yoon S, Matsuzaki Y, Mulligan RC, Melton DA. "Stemness": transcriptional profiling of embryonic and adult stem cells. Science. 2002. 298:597–600.85. Reik W. Stability and flexibility of epigenetic gene regulation in mammalian development. Nature. 2007. 447:425–432.86. Reinders J, Sickmann A. State-of-the-art in phosphoproteomics. Proteomics. 2005. 5:4052–4061.87. Rubio D, Garcia-Castro J, Martín MC, et al. Spontaneous human adult stem cell transformation. Cancer Res. 2005. 65:3035–3039.88. Russo VEA, Martienssen RA, Riggs AD. Epigenetic mechanisms of gene regulation. 1996. Plainview, N.Y.: Cold Spring Harbor Laboratory Press.89. Sanosaka T, Namihira M, Nakashima K. Epigenetic mechanisms in sequential differentiation of neural stem cells. Epigenetics. 2009. 4:89–92.90. Schwartz BE, Ahmad K. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 2005. 19:804–814.91. Shalgi R, Lieber D, Oren M, Pilpel Y. Global and Local Architecture of the Mammalian microRNA-Transcription Factor Regulatory Network. PLoS Comput Biol. 2007. 3:e131.92. Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol Syst Biol. 2007. 3:88.93. Stark A, Brennecke J, Russell RB, Cohen SM. Identification of Drosophila MicroRNA targets. PLoS Biol. 2003. 1:E60.94. Stein LD. Human genome: end of the beginning. Nature. 2004. 431:915–916.95. Stolt CC, Rehberg S, Ader M, et al. Terminal differentiation of myelin-forming oligodendrocytes depends on the transcription factor Sox10. Genes Dev. 2002. 16:165–170.96. Tai MH, Chang CC, Kiupel M, Webster JD, Olson LK, Trosko JE. Oct4 expression in adult human stem cells: evidence in support of the stem cell theory of carcinogenesis. Carcinogenesis. 2005. 26:495–502.97. Takahashi K, Tanabe K, Ohnuki M, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007. 131:861–872.98. Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006. 126:663–676.99. Taverna SD, Ueberheide BM, Liu Y, et al. Long-distance combinatorial linkage between methylation and acetylation on histone H3 N termini. Proc Natl Acad Sci U S A. 2007. 104:2086–2091.100. Tay YM, Tam WL, Ang YS, et al. MicroRNA-134 modulates the differentiation of mouse embryonic stem cells where it causes post-transcriptional attenuation of nanog and LRH1. Stem Cells. 2008. 26:17–29.101. Taylor CJ, Bolton EM, Pocock S, Sharples LD, Pedersen RA, Bradley JA. Banking on human embryonic stem cells: estimating the number of donor cell lines needed for HLA matching. Lancet. 2005. 366:2019–2025.102. The ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004. 306:636–640.103. Thomas CE, Kelleher NL, Mizzen CA. Mass spectrometric characterization of human histone H3: a bird's eye view. J Proteome Res. 2006. 5:240–247.104. Uhlen M, Bjorling E, Agaton C, et al. A human protein atlas for normal and cancer tissues based on antibody proteomics. Mol Cell Proteomics. 2005. 4:1920–1932.105. Uhlen M, Ponten F. Antibody-based proteomics for human tissue profiling. Mol Cell Proteomics. 2005. 4:384–393.106. Unwin RD, Smith DL, Blinco D, et al. Quantitative proteomics reveals posttranslational control as a regulatory factor in primary hematopoietic stem cells. Blood. 2006. 107:4687–4694.107. Van Hoof D, Muñoz J, Braam SR, et al. Phosphorylation dynamics during early differentiation of human embryonic stem cells. Cell Stem Cell. 2009. 5:214–226.108. Vogel G. Cell biology. Ready or not? Human ES cells head toward the clinic. Science. 2005. 308:1534–1538.109. Wade PA. Methyl CpG binding proteins: coupling chromatin architecture to gene regulation. Oncogene. 2001. 20:3166–3173.110. Wallin E, von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Sci. 1998. 7:1029–1038.111. Walsh C. Posttranslational modification of proteins: expanding nature's inventory. 2006. Englewood, Colo.: Roberts and Co. Publishers.112. Wang ZX, Teh CH, Chan CM, et al. The transcription factor Zfp281 controls embryonic stem cell pluripotency by direct activation and repression of target genes. Stem Cells. 2008. 26:2791–2799.113. Watt F, Molloy PL. Cytosine methylation prevents binding to DNA of a HeLa cell transcription factor required for optimal expression of the adenovirus major late promoter. Genes Dev. 1988. 2:1136–1143.114. Wenick AS, Hobert O. Genomic cis-regulatory architecture and trans-acting regulators of a single interneuron-specific gene battery in C. elegans. Dev Cell. 2004. 6:757–770.115. Wienholds E, Kloosterman WP, Miska E, et al. MicroRNA expression in zebrafish embryonic development. Science. 2005. 309:310–311.116. Wienholds E, Plasterk RH. MicroRNA function in animal development. FEBS Lett. 2005. 579:5911–5922.117. Wissmuller S, Kosian T, Wolf M, Finzsch M, Wegner M. The high-mobility-group domain of Sox proteins interacts with DNA-binding domains of many transcription factors. Nucleic Acids Res. 2006. 34:1735–1744.118. Woolfson A, Ellmark P, Chrisp JS, A Scott M, Christopherson RI. The application of CD antigen proteomics to pharmacogenomics. Pharmacogenomics. 2006. 7:759–771.119. Wu CC, MacCoss MJ, Howell KE, Yates JR 3rd. A method for the comprehensive proteomic analysis of membrane proteins. Nat Biotechnol. 2003. 21:532–538.120. Wu CC, Yates JR 3rd. The application of mass spectrometry to membrane proteomics. Nat Biotechnol. 2003. 21:262–267.121. Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007. 131:146–159.122. Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004. 20:617–624.123. Yoo AS, Greenwald I. LIN-12/Notch activation leads to microRNA-mediated down-regulation of Vav in C. elegans. Science. 2005. 310:1330–1333.124. Yu J, Vodyanik MA, Smuga-Otto K, et al. Induced Pluripotent Stem Cell Lines Derived from Human Somatic Cells. Science. 2007. 318:1917–1920.125. Yu L, Gaskell SJ, Brookman JL. Epitope mapping of monoclonal antibodies by mass spectrometry: identification of protein antigens in complex biological systems. J Am Soc Mass Spectrom. 1998. 9:208–215.126. Zola H, Swart B, Nicholson I, et al. CD molecules 2005: human cell differentiation molecules. Blood. 2005. 106:3123–3126.127. Zola H, Swart BW. Human leucocyte differentiation antigens. Trends Immunol. 2003. 24:353–354.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Induced pluripotent stem cells and personalized medicine: current progress and future perspectives
- Data Mining for High Dimensional Data in Drug Discovery and Development
- Mass spectrometry based proteomic analysis of human stem cells: a brief review
- Systems Bioinformatics Research Trends
- Human Genome Project