Cancer Res Treat.
2004 Feb;36(1):31-42.
Expression Profiling of the Cellular Processes in Uterine Leiomyomas: Omic Approaches and IGF-2 Association with Leiomyosarcomas
- Affiliations
-
- 1Catholic Research Institutes of Medical Science, College of Medicine, The Catholic University of Korea, Korea.
- 2Department of Obstetrics and Gynecology, College of Medicine, The Catholic University of Korea, Korea.
- 3College of Pharmacy, Seoul National University, Seoul, Korea.
Abstract
- PURPOSE
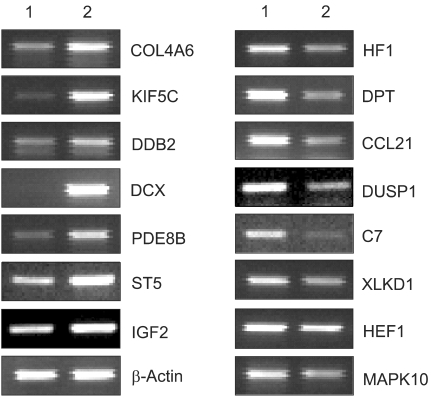
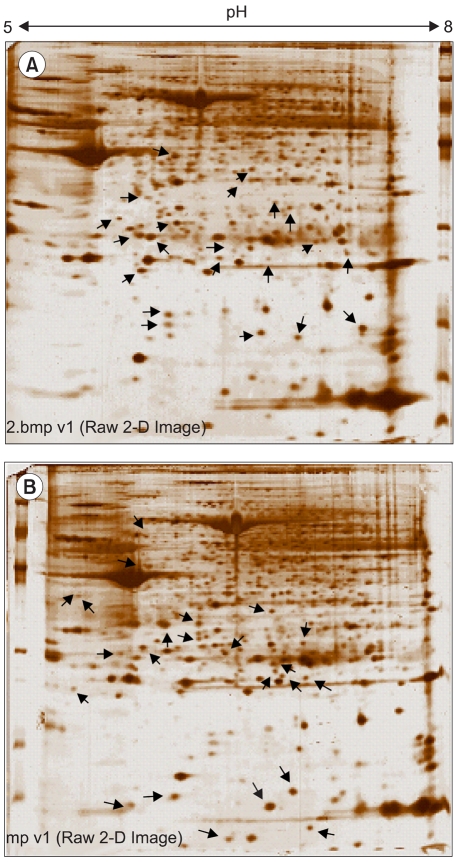
This study utilized both cDNA microarray and 2D protein gel electrophoresis technology to investigate the multiple interactions of the genes and proteins involved in the pathophysiology of uterine leiomyomas. Also, Gene Ontology (GO) analysis was used to systematically characterize the global expression profiles, which were found to correlate with the leiomyosarcomas. MATERIALS AND METHODS: The uterine leiomyoma biopsies were obtained from patients in the Department of Obstetrics and Gynecology, The Catholic University of Korea. Differentially expressed transcriptome and proteome, in 6 paired leiomyoma and normal myometrium, were profiled. The total RNAs from the leiomyoma and normal myometrium were labeled with Cy5 and Cy3. All specimens were punch-biopsy-obtained, and frozen in liquid nitrogen. RESULTS: Screening of up to 17, 000 genes identified 71 that were either up-regulated or down-regulated (21 and 50, respectively). The gene expression profiles were classified into 420 mutually dependent functional sets, resulting in 611 cellular processes, according to the gene ontology. Also, the protein analysis, using 2D gel electrophoresis, identified 33 proteins (17 up-regulated and 16 down-regulated) with more than 500 total spots, which were classified into 302 cellular processes. Of these functional profilings, transcriptomes and proteoms down- regulations were shown in the cell adhesion, cell motility, organogenesis, enzyme regulator, structural molecule activity and responses to external stimulus functional activities, which are supposed to play important roles in the pathophysiology. In contrast, up-regulation was only shown in the nucleic acid binding activity. The CDKN2A, ADH1A, DCX, IGF2, CRABP2 and KIF5C were found to increase the reliability of this study, and correlate with the leiomyosarcomas. CONCLUSION: Potentially significant pathogenetic cellular processes showed that down-regulated functional profiling has an important impact on the discovery of the pathogenic pathways in leiomyomas and leiomyosarcomas. GO analysis can also overcome the complexity of the expression profiles of cDNA microarrays and 2D protein analyses, via a cellular process level approach. Thereby, a valuable prognostic candidate gene, with real relevance to disease-specific pathogenesis, can be found at cellular process levels.
MeSH Terms
-
Animals
Biopsy
Cell Adhesion
Cell Movement
Electrophoresis
Electrophoresis, Gel, Two-Dimensional
Female
Gene Ontology
Gynecology
Humans
Insulin-Like Growth Factor II*
Korea
Leiomyoma*
Leiomyosarcoma*
Mass Screening
Mice
Myometrium
Nitrogen
Obstetrics
Oligonucleotide Array Sequence Analysis
Organogenesis
Proteome
Proteomics
RNA
Social Control, Formal
Transcriptome
Up-Regulation
Insulin-Like Growth Factor II
Nitrogen
Proteome
RNA
Figure
Reference
-
1. Buttram VC Jr, Reiter RC. Uterine leiomyomata: etiology, symptomatology, and management. Fertil Steril. 1981; 36:433–445. PMID: 7026295.2. Ligon AH, Morton CC. Leiomyomata: heritability and cytogenetic studies. Hum Reprod Update. 2001; 7:8–14. PMID: 11212080.
Article3. McKeeby JL, Li X, Zhuang Z, Vortmeyer AO, Huang S, Pirner M, Skarulis MC, James-Newton L, Marx SJ, Lubensky IA. Multiple leiomyomas of the esophagus, lung, and uterus in multiple endocrine neoplasia type 1. Am J Pathol. 2001; 159:1121–1127. PMID: 11549605.
Article4. Vu K, Greenspan DL, Wu TC, Zacur HA, Kurman RJ. Cellular proliferation, estrogen receptor, progesterone receptor, and bcl-2 expres-sion in GnRH agonist-treated uterine leiomyomas. Hum Pathol. 1998; 29:359–363. PMID: 9563785.
Article5. Edmonson JH, Blessing JA, Cosin JA, Miller DS, Cohn DE, Rotmensch J. Phase II study of mitomycin, doxorubicin, and cisplatin in the treatment of advanced uterine leiomyosarcoma: a Gynecologic Oncology Group study. Gynecol Oncol. 2002; 85:507–510. PMID: 12051882.
Article6. Kim YW, Suh MJ, Bae JS, Bae SM, Yoon JH, Hur SY, Kim JH, Ro DY, Lee JM, Namkoong SE, Kim CK, Ahn WS. New approaches to functional process discovery in HPV 16-associated cervical cancer cells by gene ontology. Cancer Res Treat. 2003; 35:304–313.
Article7. Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using gene ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003; 4:R7–R12. PMID: 12540299.8. Tsibris JC, Segars J, Coppola D, Mane S, Wilbanks GD, O'Brien WF, Spellacy WN. Insights from gene arrays on the development and growth regulation of uterine leiomyomata. Fertil Steril. 2002; 78:114–121. PMID: 12095500.
Article9. Eisen MB, Brown PO. DNA arrays for analysis of gene expression. Methods Enzymol. 1999; 303:179–205. PMID: 10349646.10. DeRisi JL, Iyer VR, Brown PO. Exploring the metabolic and genetic control of gene expression on a genomic scale. Science. 1997; 278:680–686. PMID: 9381177.
Article11. Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Nat Acad Sci USA. 1998; 95:14863–14868. PMID: 9843981.
Article12. Pletcher SD, Macdonald SJ, Marguerie R, Certa U, Stearns SC, Goldstein DB, Partridge L. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Curr Biol. 2002; 12:712–723. PMID: 12007414.
Article13. Juan HF, Lin JY, Chang WH, Wu CY, Pan TL, Tseng MJ, Khoo KH, Chen ST. Biomic study of human myeloid leukemia cells differentiation to macrophages using DNA array, proteomic, and bioinformatic analytical methods. Electrophoresis. 2002; 23:2490–2504. PMID: 12210208.
Article14. Skubitz KM, Skubitz AP. Differential gene expression in uterine leiomyoma. J Lab Clin Med. 2003; 141:297–308. PMID: 12761473.
Article15. Brinkmann U, Vasmatzis G, Lee B, Yerushalmi N, Essand M, Pastan I. PAGE-1, an X chromosome-linked GAGE-like gene that is expressed in normal and neoplastic prostate, testis, and uterus. Proc Nat Acad Sci USA. 1998; 95:10757–10762. PMID: 9724777.
Article16. Chegini N, Verala J, Luo X, Xu J, Williams RS. Gene expression profile of leiomyoma and myometrium and the effect of gonadotropin releasing hormone analogue therapy. J Soc Gynecol Investig. 2003; 10:161–171.
Article17. Ogilvie P, Bardi G, Clark-Lewis I, Baggiolini M, Uguccioni M. Eotaxin is a natural antagonist for CCR2 and an agonist for CCR5. Blood. 2001; 97:1920–1924. PMID: 11264152.
Article18. Sugimoto M, Oohashi T, Ninomiya Y. The genes COL4A5 and COL4A6, coding for basement membrane collagen chains alpha-5 (IV) and alpha-6 (IV) are located head-to-head in close proximity on human chromosome Xq22 and COL4A6 is transcribed from two alternative promoters. Proc Nat Acad Sci USA. 1994; 91:11679–11683. PMID: 7972123.19. Walther A, Riehemann K, Gerke V. A novel ligand of the formyl peptide receptor: annexin I regulates neutrophil extravasation by interacting with the FPR. Mol Cell. 2000; 5:831–840. PMID: 10882119.20. Banerji S, Ni J, Wang SX, Clasper S, Su J, Tammi R, Jones M, Jackson DG. LYVE-1, a new homologue of the CD44 glycoprotein, is a lymph-specific receptor for hyaluronan. J Cell Biol. 1999; 144:789–801. PMID: 10037799.
Article21. Zhu D, Kepley CL, Zhang M, Zhang K, Saxon A. A novel human immunoglobulin Fc-gamma-Fc-epsilon bifunctional fusion protein inhibits Fc-epsilon-RI-mediated degranulation. Nature Med. 2002; 8:518–521. PMID: 11984598.22. Egan LJ, Orren A, Doherty J, Wurzner R, McCarthy CF. Hereditary deficiency of the seventh component of complement and recurrent meningococcal infection: investigations of an Irish family using a novel haemolytic screening assay for complement activity and C7 M/N allotyping. Epidemiol Infect. 1994; 113:275–281. PMID: 7523157.
Article23. Skubitz KM, Skubitz AP. Differential gene expression in leiomyosarcoma. Cancer. 2003; 98:1029–1038. PMID: 12942572.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Expressions and Diagnostic Usefulness of MIB-1 and p53 in Uterine Smooth Muscle Tumors
- Current Medical Therapy for Uterine Leiomyomas
- Expression of insulin-like growth factor [IGF] -Ia , IGF-Ib , and IGF-II messenger ribonucleic acid in uterine leiomyoma
- Correlation between Expression of Insulin-like Growth Factor-I Receptor and Clinicopathologic Prognostic Factors in Invasive Ductal Carcinoma of the Breast
- Degenerated Uterine Leiomyomas Mimicking Malignant Bilateral Ovarian Surface Epithelial Tumors