Brain Tumor Res Treat.
2014 Apr;2(1):1-6. 10.14791/btrt.2014.2.1.1.
Roles of Long Non-Coding RNAs on Tumorigenesis and Glioma Development
- Affiliations
-
- 1Cancer Cell and Molecular Biology Branch, Research Institute, National Cancer Center, Goyang, Korea. jhkim@ncc.re.kr
- 2Specific Organs Cancer Branch, Research Institute, National Cancer Center, Goyang, Korea.
- 3Department of System Cancer Science, Graduate School of Cancer Science and Policy, National Cancer Center, Goyang, Korea.
- KMID: 1734662
- DOI: http://doi.org/10.14791/btrt.2014.2.1.1
Abstract
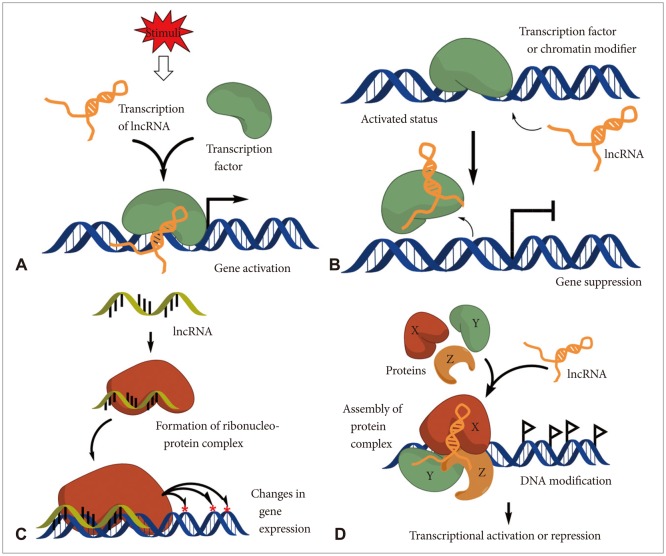
- More than 98% of eukaryotic transcriptomes are composed of non-coding RNAs with no functional protein-coding capacity. Those transcripts also include tens of thousands of long non-coding RNAs (lncRNAs) which are emerging as key elements of cellular homeostasis, essentially tumorigenesis steps. However, we are only beginning to understand the nature and extent of the involvement of lncRNAs on tumorigeneis. Here, we highlight recent progresses that have identified a myriad of molecular functions on tumorigenesis for several lncRNAs including metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), prostate cancer associated non-coding RNA 1 (PRNCR1), prostate cancer gene expression marker 1 (PCGEM1), H19, and homeobox transcript antisense intergenic RNA (HOTAIR), and several new lncRNAs for glioma development. Potential therapeutic approaches for the lncRNAs in various human diseases are also discussed.
Keyword
MeSH Terms
Figure
Reference
-
1. Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005; 309:1559–1563. PMID: 16141072.2. International Human Genome Sequencing Consortium. Finishing the euchromatic sequence of the human genome. Nature. 2004; 431:931–945. PMID: 15496913.3. Esteller M. Non-coding RNAs in human disease. Nat Rev Genet. 2011; 12:861–874. PMID: 22094949.
Article4. Kapranov P, Cheng J, Dike S, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007; 316:1484–1488. PMID: 17510325.
Article5. Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007; 8:413–423. PMID: 17486121.
Article6. Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006; 6:259–269. PMID: 16557279.
Article7. Bartolomei MS, Zemel S, Tilghman SM. Parental imprinting of the mouse H19 gene. Nature. 1991; 351:153–155. PMID: 1709450.
Article8. Qiu MT, Hu JW, Yin R, Xu L. Long noncoding RNA: an emerging paradigm of cancer research. Tumour Biol. 2013; 34:613–620. PMID: 23359273.
Article9. Gibb EA, Vucic EA, Enfield KS, et al. Human cancer long non-coding RNA transcriptomes. PLoS One. 2011; 6:e25915. PMID: 21991387.
Article10. Hung T, Chang HY. Long noncoding RNA in genome regulation: prospects and mechanisms. RNA Biol. 2010; 7:582–585. PMID: 20930520.11. Shi X, Sun M, Liu H, Yao Y, Song Y. Long non-coding RNAs: a new frontier in the study of human diseases. Cancer Lett. 2013; 339:159–166. PMID: 23791884.
Article12. Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011; 43:904–914. PMID: 21925379.
Article13. Kallen AN, Zhou XB, Xu J, et al. The imprinted H19 lncRNA antago nizes let-7 microRNAs. Mol Cell. 2013; 52:101–112. PMID: 24055342.14. Yang L, Lin C, Jin C, et al. lncRNA-dependent mechanisms of androgen-receptor-regulated gene activation programs. Nature. 2013; 500:598–602. PMID: 23945587.
Article15. Plath K, Fang J, Mlynarczyk-Evans SK, et al. Role of histone H3 lysine 27 methylation in X inactivation. Science. 2003; 300:131–135. PMID: 12649488.
Article16. Huarte M, Guttman M, Feldser D, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010; 142:409–419. PMID: 20673990.
Article17. Bejerano G, Pheasant M, Makunin I, et al. Ultraconserved elements in the human genome. Science. 2004; 304:1321–1325. PMID: 15131266.
Article18. Lujambio A, Portela A, Liz J, et al. CpG island hypermethylation-associated silencing of non-coding RNAs transcribed from ultraconserved regions in human cancer. Oncogene. 2010; 29:6390–6401. PMID: 20802525.
Article19. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. 2012; 81:145–166. PMID: 22663078.
Article20. Gupta RA, Shah N, Wang KC, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010; 464:1071–1076. PMID: 20393566.
Article21. Ji P, Diederichs S, Wang W, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003; 22:8031–8041. PMID: 12970751.22. Wilusz JE, Freier SM, Spector DL. 3' end processing of a long nuclear-retained noncoding RNA yields a tRNA-like cytoplasmic RNA. Cell. 2008; 135:919–932. PMID: 19041754.
Article23. Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010; 39:925–938. PMID: 20797886.
Article24. Jurica MS, Moore MJ. Pre-mRNA splicing: awash in a sea of proteins. Mol Cell. 2003; 12:5–14. PMID: 12887888.25. Manley JL, Krainer AR. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev. 2010; 24:1073–1074. PMID: 20516191.26. Gutschner T, Hämmerle M, Eissmann M, et al. The noncoding RNA MALAT1 is a critical regulator of the metastasis phenotype of lung cancer cells. Cancer Res. 2013; 73:1180–1189. PMID: 23243023.
Article27. Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013; 9:e1003368. PMID: 23555285.
Article28. Joaquin M, Watson RJ. Cell cycle regulation by the B-Myb transcription factor. Cell Mol Life Sci. 2003; 60:2389–2401. PMID: 14625684.
Article29. Petrovics G, Zhang W, Makarem M, et al. Elevated expression of PCGEM1, a prostate-specific gene with cell growth-promoting function, is associated with high-risk prostate cancer patients. Oncogene. 2004; 23:605–611. PMID: 14724589.
Article30. Chung S, Nakagawa H, Uemura M, et al. Association of a novel long non-coding RNA in 8q24 with prostate cancer susceptibility. Cancer Sci. 2011; 102:245–252. PMID: 20874843.
Article31. Komiya A, Yasuda K, Watanabe A, Fujiuchi Y, Tsuzuki T, Fuse H. The prognostic significance of loss of the androgen receptor and neuroendocrine differentiation in prostate biopsy specimens among castration-resistant prostate cancer patients. Mol Clin Oncol. 2013; 1:257–262. PMID: 24649157.
Article32. Brannan CI, Dees EC, Ingram RS, Tilghman SM. The product of the H19 gene may function as an RNA. Mol Cell Biol. 1990; 10:28–36. PMID: 1688465.
Article33. Rachmilewitz J, Goshen R, Ariel I, Schneider T, de Groot N, Hochberg A. Parental imprinting of the human H19 gene. FEBS Lett. 1992; 309:25–28. PMID: 1380925.
Article34. Keniry A, Oxley D, Monnier P, et al. The H19 lincRNA is a developmental reservoir of miR-675 that suppresses growth and Igf1r. Nat Cell Biol. 2012; 14:659–665. PMID: 22684254.
Article35. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Long non-coding RNA H19 increases bladder cancer metastasis by associating with EZH2 and inhibiting E-cadherin expression. Cancer Lett. 2013; 333:213–221. PMID: 23354591.
Article36. Luo M, Li Z, Wang W, Zeng Y, Liu Z, Qiu J. Upregulated H19 contributes to bladder cancer cell proliferation by regulating ID2 expression. FEBS J. 2013; 280:1709–1716. PMID: 23399020.
Article37. Matouk IJ, DeGroot N, Mezan S, et al. The H19 non-coding RNA is essential for human tumor growth. PLoS One. 2007; 2:e845. PMID: 17786216.
Article38. Matouk IJ, Mezan S, Mizrahi A, et al. The oncofetal H19 RNA connection: hypoxia, p53 and cancer. Biochim Biophys Acta. 2010; 1803:443–451. PMID: 20117150.
Article39. Tsang WP, Ng EK, Ng SS, et al. Oncofetal H19-derived miR-675 regulates tumor suppressor RB in human colorectal cancer. Carcinogenesis. 2010; 31:350–358. PMID: 19926638.
Article40. Zhang L, Yang F, Yuan JH, et al. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis. 2013; 34:577–586. PMID: 23222811.
Article41. Cai X, Cullen BR. The imprinted H19 noncoding RNA is a primary microRNA precursor. RNA. 2007; 13:313–316. PMID: 17237358.
Article42. Gwak HS, Kim TH, Jo GH, et al. Silencing of microRNA-21 confers radio-sensitivity through inhibition of the PI3K/AKT pathway and enhancing autophagy in malignant glioma cell lines. PLoS One. 2012; 7:e47449. PMID: 23077620.
Article43. Kwak HJ, Kim YJ, Chun KR, et al. Downregulation of Spry2 by miR-21 triggers malignancy in human gliomas. Oncogene. 2011; 30:2433–2442. PMID: 21278789.
Article44. Kim YJ, Park SJ, Choi EY, et al. PTEN modulates miR-21 processing via RNA-regulatory protein RNH1. PLoS One. 2011; 6:e28308. PMID: 22162762.
Article45. Ellis BC, Molloy PL, Graham LD. CRNDE: A Long Non-Coding RNA Involved in CanceR, Neurobiology, and DEvelopment. Front Genet. 2012; 3:270. PMID: 23226159.
Article46. Graham LD, Pedersen SK, Brown GS, et al. Colorectal Neoplasia Differentially Expressed (CRNDE), a Novel Gene with Elevated Expression in Colorectal Adenomas and Adenocarcinomas. Genes Cancer. 2011; 2:829–840. PMID: 22393467.
Article47. Wang P, Ren Z, Sun P. Overexpression of the long non-coding RNA MEG3 impairs in vitro glioma cell proliferation. J Cell Biochem. 2012; 113:1868–1874. PMID: 22234798.
Article48. Shi Y, Wang Y, Luan W, et al. Long Non-Coding RNA H19 Promotes Glioma Cell Invasion by Deriving miR-675. PLoS One. 2014; 9:e86295. PMID: 24466011.
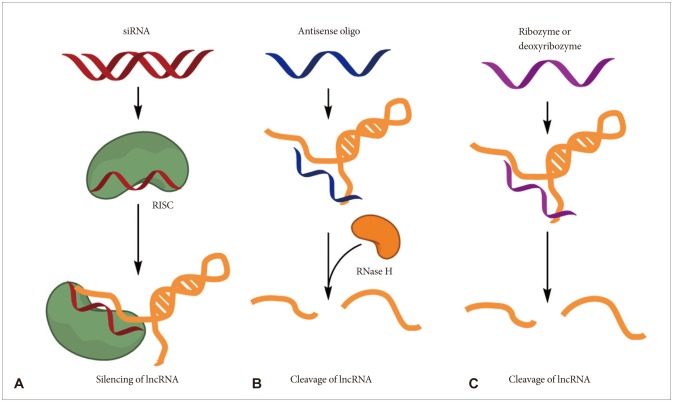
Article49. Li CH, Chen Y. Targeting long non-coding RNAs in cancers: progress and prospects. Int J Biochem Cell Biol. 2013; 45:1895–1910. PMID: 23748105.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Deciphering the Role of Non-Coding RNAs as Regulators in the Wound Healing Process
- Roles of Oncogenic Long Non-coding RNAs in Cancer Development
- Role of Long Non-coding Ribonucleic Acid in Gastrointestinal Cancer
- Application of Long Non-coding RNAs in Gastric Cancer
- Long Noncoding RNA PVT1 Promotes Stemness and Temozolomide Resistance through miR-365/ELF4/SOX2 Axis in Glioma