Cancer Res Treat.
2023 Jan;55(1):145-154. 10.4143/crt.2022.001.
Analysis of PIK3CA Mutation Concordance and Frequency in Primary and Different Distant Metastatic Sites in Breast Cancer
- Affiliations
-
- 1Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Korea
- 2Department of Pathology and Translational Genomics, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 3Department of Health Sciences and Technology, SAIHST, Sungkyunkwan University, Seoul, Korea
- 4Laboratory of Cancer Genomics and Molecular Pathology, Samsung Medical Center, Sungkyunkwan University School of Medicine, Seoul, Korea
- 5Central Laboratory, LOGONE Bio-Convergence Research Foundation, Seoul, Korea
- 6Laboratory of Molecular Pathology and Cancer Genomics, College of Pharmacy and Research Institute of Pharmaceutical Sciences, Seoul National University, Seoul, Korea
- KMID: 2538000
- DOI: http://doi.org/10.4143/crt.2022.001
Abstract
- Purpose
The purpose of this study was to investigate the concordance rate of PIK3CA mutations between primary and matched distant metastatic sites in patients with breast cancer and to verify whether there are differences in the frequency of PIK3CA hotspot mutations depending on the metastatic sites involved.
Materials and Methods
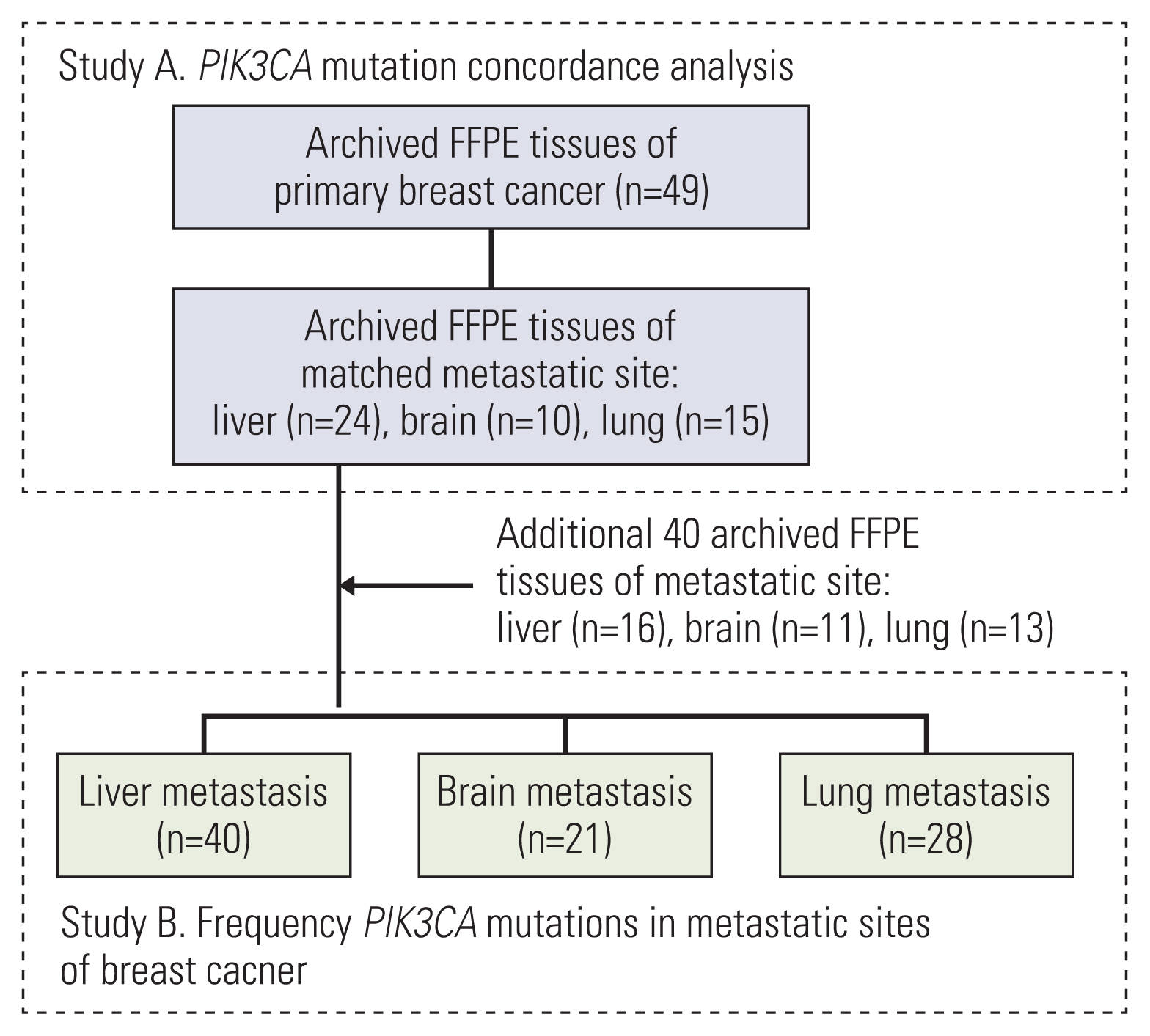
Archived formalin-fixed paraffin-embedded (FFPE) specimens of primary breast and matched distant metastatic tumors were retrospectively obtained for 49 patients. Additionally, 40 archived FFPE specimens were independently collected from different breast cancer metastatic sites, which were limited to three common sites: the liver, brain, and lung. PIK3CA mutations were analyzed using droplet digital PCR, including hotspots involving exons 9 and 20.
Results
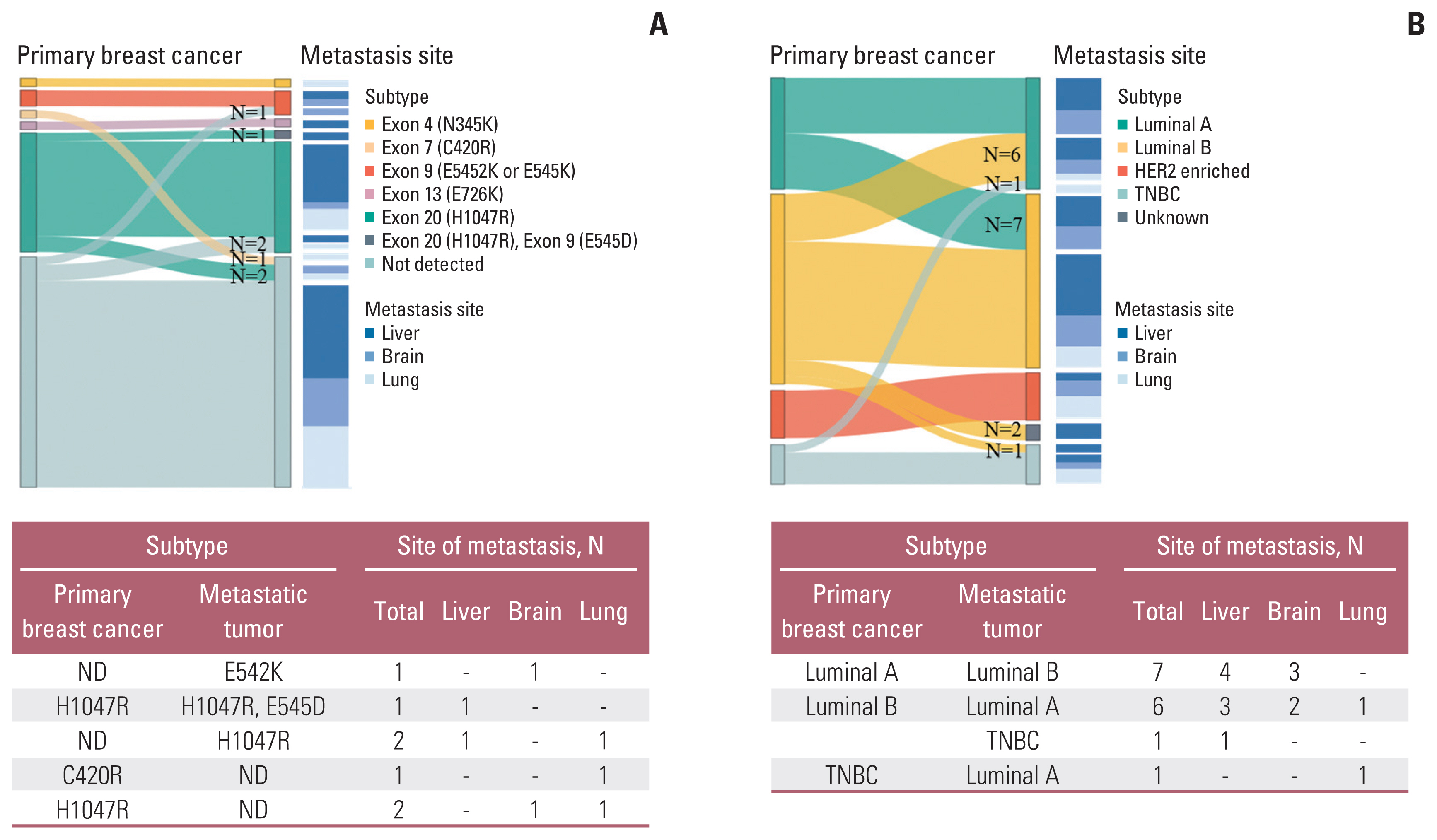
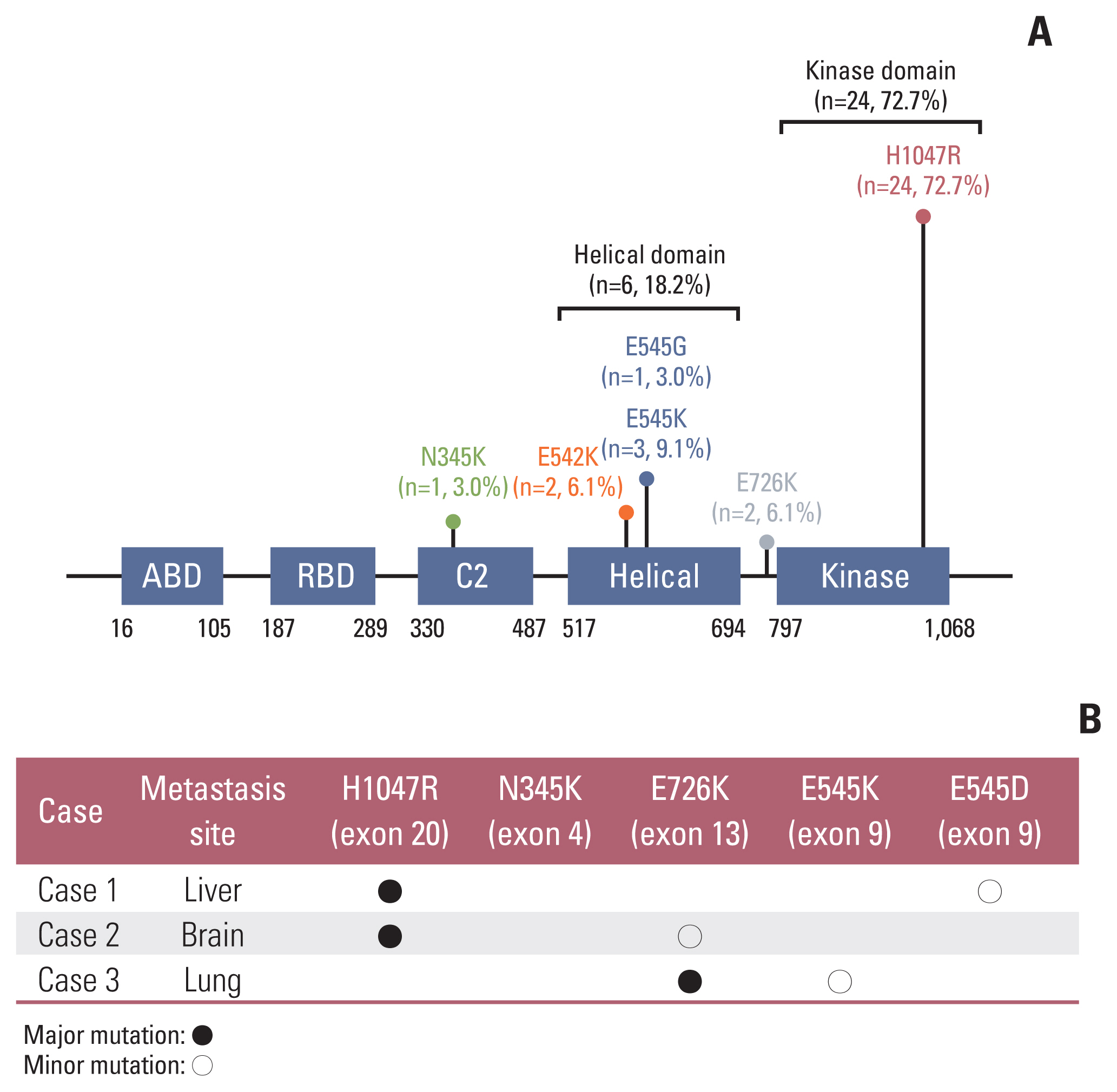
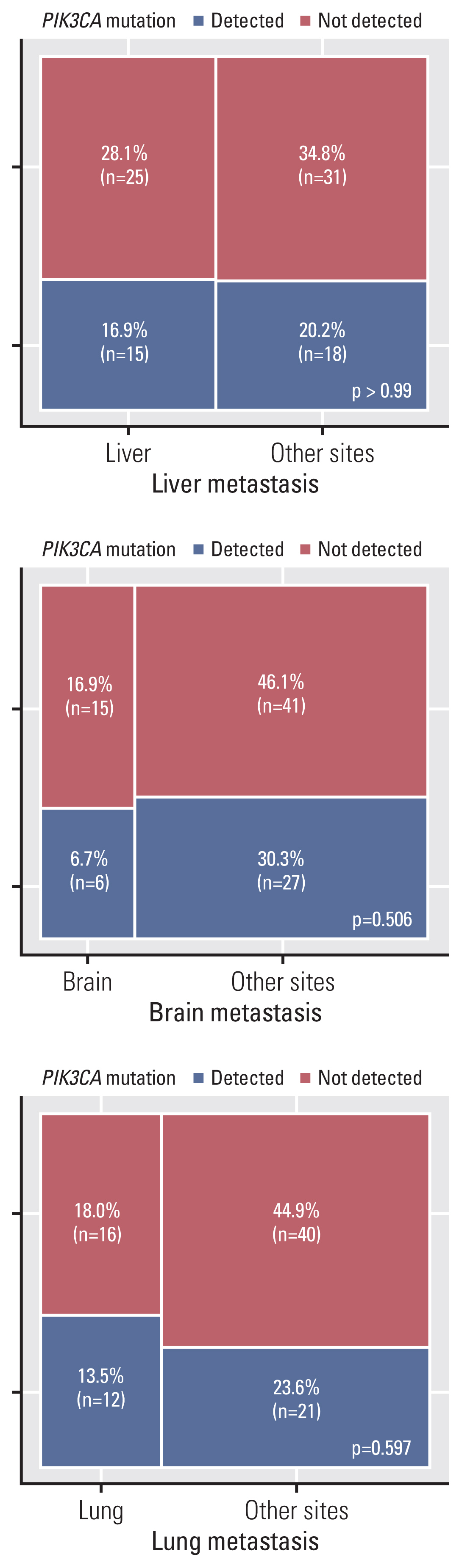
After analysis of 49 breast tumors with matched metastasis sites, 87.8% showed concordance in PIK3CA mutation status. According to PIK3CA hotspot mutation testing in 89 cases of breast cancer metastatic sites, the proportion of PIK3CA mutations at sites of metastasis involving the liver, brain, and lung was 37.5%, 28.6%, and 42.9%, respectively, which did not result in statistical significance.
Conclusion
The high concordance of PIK3CA mutation status between primary and matched metastasis sites suggests that metastatic sites, regardless of the metastatic organ, could be considered sample sources for PIK3CA mutation testing for improved therapeutic strategies in patients with metastatic breast cancer.
Figure
Reference
-
References
1. Pan H, Gray R, Braybrooke J, Davies C, Taylor C, McGale P, et al. 20-Year risks of breast-cancer recurrence after stopping endocrine therapy at 5 years. N Engl J Med. 2017; 377:1836–46.
Article2. Gradishar WJ, Anderson BO, Abraham J, Aft R, Agnese D, Allison KH, et al. Breast cancer, version 3.2020, NCCN Clinical Practice Guidelines in Oncology. J Natl Compr Canc Netw. 2020; 18:452–78.3. Janku F, Yap TA, Meric-Bernstam F. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol. 2018; 15:273–91.
Article4. Karakas B, Bachman KE, Park BH. Mutation of the PIK3CA oncogene in human cancers. Br J Cancer. 2006; 94:455–9.
Article5. Ligresti G, Militello L, Steelman LS, Cavallaro A, Basile F, Nicoletti F, et al. PIK3CA mutations in human solid tumors: role in sensitivity to various therapeutic approaches. Cell Cycle. 2009; 8:1352–8.
Article6. Isakoff SJ, Engelman JA, Irie HY, Luo J, Brachmann SM, Pearline RV, et al. Breast cancer-associated PIK3CA mutations are oncogenic in mammary epithelial cells. Cancer Res. 2005; 65:10992–1000.7. Mosele F, Stefanovska B, Lusque A, Tran Dien A, Garberis I, Droin N, et al. Outcome and molecular landscape of patients with PIK3CA-mutated metastatic breast cancer. Ann Oncol. 2020; 31:377–86.
Article8. Anderson EJ, Mollon LE, Dean JL, Warholak TL, Aizer A, Platt EA, et al. A systematic review of the prevalence and diagnostic workup of PIK3CA mutations in HR+/HER2− metastatic breast cancer. Int J Breast Cancer. 2020; 2020:3759179.
Article9. Deng L, Zhu X, Sun Y, Wang J, Zhong X, Li J, et al. Prevalence and prognostic role of PIK3CA/AKT1 mutations in Chinese breast cancer patients. Cancer Res Treat. 2019; 51:128–40.
Article10. Vuylsteke P, Huizing M, Petrakova K, Roylance R, Laing R, Chan S, et al. Pictilisib PI3Kinase inhibitor (a phosphatidylinositol 3-kinase [PI3K] inhibitor) plus paclitaxel for the treatment of hormone receptor-positive, HER2-negative, locally recurrent, or metastatic breast cancer: interim analysis of the multicentre, placebo-controlled, phase II randomised PEGGY study. Ann Oncol. 2016; 27:2059–66.
Article11. Krop IE, Mayer IA, Ganju V, Dickler M, Johnston S, Morales S, et al. Pictilisib for oestrogen receptor-positive, aromatase inhibitor-resistant, advanced or metastatic breast cancer (FERGI): a randomised, double-blind, placebo-controlled, phase 2 trial. Lancet Oncol. 2016; 17:811–21.
Article12. Baselga J, Im SA, Iwata H, Cortes J, De Laurentiis M, Jiang Z, et al. Buparlisib plus fulvestrant versus placebo plus fulvestrant in postmenopausal, hormone receptor-positive, HER2-negative, advanced breast cancer (BELLE-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017; 18:904–16.
Article13. Di Leo A, Johnston S, Lee KS, Ciruelos E, Lonning PE, Janni W, et al. Buparlisib plus fulvestrant in postmenopausal women with hormone-receptor-positive, HER2-negative, advanced breast cancer progressing on or after mTOR inhibition (BELLE-3): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2018; 19:87–100.
Article14. Yang J, Nie J, Ma X, Wei Y, Peng Y, Wei X. Targeting PI3K in cancer: mechanisms and advances in clinical trials. Mol Cancer. 2019; 18:26.
Article15. Andre F, Ciruelos EM, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib (ALP)+fulvestrant (FUL) for advanced breast cancer (ABC): results of the phase III SOLAR-1 trial. Ann Oncol. 2018; 29(Suppl 8):VIII709.16. Allison KH, Hammond MEH, Dowsett M, McKernin SE, Cary LA, Fitzgibbons PL, et al. Estrogen and progesterone receptor testing in breast cancer: ASCO/CAP guideline update. J Clin Oncol. 2020; 38:1346–66.
Article17. Goldhirsch A, Winer EP, Coates AS, Gelber RD, Piccart-Gebhart M, Thurlimann B, et al. Personalizing the treatment of women with early breast cancer: highlights of the St Gallen International Expert Consensus on the Primary Therapy of Early Breast Cancer 2013. Ann Oncol. 2013; 24:2206–23.
Article18. Andre F, Ciruelos E, Rubovszky G, Campone M, Loibl S, Rugo HS, et al. Alpelisib for PIK3CA-mutated, hormone receptor-positive advanced breast cancer. N Engl J Med. 2019; 380:1929–40.19. Cizkova M, Susini A, Vacher S, Cizeron-Clairac G, Andrieu C, Driouch K, et al. PIK3CA mutation impact on survival in breast cancer patients and in ERalpha, PR and ERBB2-based subgroups. Breast Cancer Res. 2012; 14:R28.20. Campbell IG, Russell SE, Choong DY, Montgomery KG, Ciavarella ML, Hooi CS, et al. Mutation of the PIK3CA gene in ovarian and breast cancer. Cancer Res. 2004; 64:7678–81.21. U.S. Food and Drug Administration. Summary of safety and effectiveness data of therascreen PIK3CA RGQ PCR kit [Internet]. Silver Spring MD: U.S. Food and Drug Administration;2019. [cited 2022 Jan 2]. Available from: https://www.accessdata.fda.gov/scripts/cdrh/cfdocs/cfpma/pma.cfm?id=P190004.22. Zhu G, Ye X, Dong Z, Lu YC, Sun Y, Liu Y, et al. Highly sensitive droplet digital PCR method for detection of EGFR-activating mutations in plasma cell-free DNA from patients with advanced non-small cell lung cancer. J Mol Diagn. 2015; 17:265–72.
Article23. Olmedillas-Lopez S, Garcia-Arranz M, Garcia-Olmo D. Current and emerging applications of droplet digital PCR in oncology. Mol Diagn Ther. 2017; 21:493–510.
Article24. Taylor SC, Laperriere G, Germain H. Droplet Digital PCR versus qPCR for gene expression analysis with low abundant targets: from variable nonsense to publication quality data. Sci Rep. 2017; 7:2409.25. Kim SS, Choi HJ, Kim JJ, Kim MS, Lee IS, Byun B, et al. Droplet digital PCR-based EGFR mutation detection with an internal quality control index to determine the quality of DNA. Sci Rep. 2018; 8:543.
Article26. Kim S, Park C, Ji Y, Kim DG, Bae H, van Vrancken M, et al. Deamination effects in formalin-fixed, paraffin-embedded tissue samples in the era of precision medicine. J Mol Diagn. 2017; 19:137–46.27. Do H, Dobrovic A. Sequence artifacts in DNA from formalin-fixed tissues: causes and strategies for minimization. Clin Chem. 2015; 61:64–71.
Article28. Vasan N, Razavi P, Johnson JL, Shao H, Shah H, Antoine A, et al. Double PIK3CA mutations in cis increase oncogenicity and sensitivity to PI3Kalpha inhibitors. Science. 2019; 366:714–23.
Article29. Martinez-Saez O, Chic N, Pascual T, Adamo B, Vidal M, Gonzalez-Farre B, et al. Frequency and spectrum of PIK3CA somatic mutations in breast cancer. Breast Cancer Res. 2020; 22:45.30. Pierobon M, Ramos C, Wong S, Hodge KA, Aldrich J, Byron S, et al. Enrichment of PI3K-AKT-mTOR pathway activation in hepatic metastases from breast cancer. Clin Cancer Res. 2017; 23:4919–28.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Prevalence and Prognostic Role of PIK3CA/AKT1 Mutations in Chinese Breast Cancer Patients
- PIK3CA H1047R Mutation Associated with a Lower Pathological Complete Response Rate in Triple-Negative Breast Cancer Patients Treated with Anthracycline-Taxane–Based Neoadjuvant Chemotherapy
- Correlation between p53 and MIB1 Index Expression of Primary Tumor and Metastatic Lymph Node in Breast Cancer
- The p53 Mutation and DNA Ploidy in Human Metastatic Breast Cancer
- Concomitant PIK3CA and TP53 Mutations in Breast Cancer: An Analysis of Clinicopathologic and Mutational Features, Neoadjuvant Therapeutic Response, and Prognosis