J Pathol Transl Med.
2020 Sep;54(5):378-386. 10.4132/jptm.2020.06.01.
Prediction of TP53 mutations by p53 immunohistochemistry and their prognostic significance in gastric cancer
- Affiliations
-
- 1Department of Pathology, Seoul National University Bundang Hospital, Seoul National University College of Medicine, Seongnam, Korea
- 2Department of Pathology, Gangnam Severance Hospital, Yonsei University College of Medicine, Seoul, Korea
- 3Department of Pathology, Seoul National University Hospital, Seoul National University College of Medicine, Seoul, Korea
- KMID: 2506271
- DOI: http://doi.org/10.4132/jptm.2020.06.01
Abstract
- Background
Recently, molecular classifications of gastric cancer (GC) have been proposed that include TP53 mutations and their functional activity. We aimed to demonstrate the correlation between p53 immunohistochemistry (IHC) and TP53 mutations as well as their clinicopathological significance in GC.
Methods
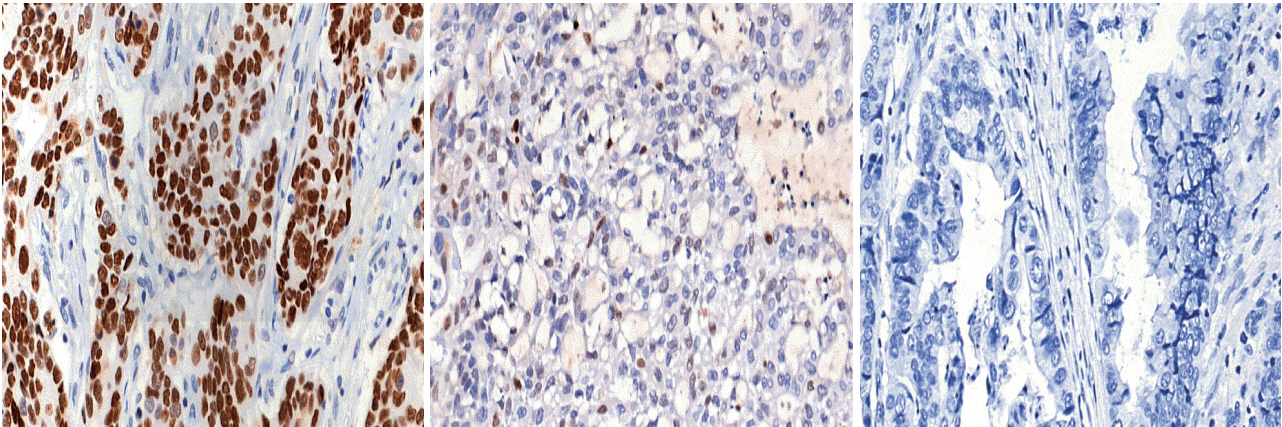
Deep targeted sequencing was performed using surgical or biopsy specimens from 120 patients with GC. IHC for p53 was performed and interpreted as strong, weak, or negative expression. In 18 cases (15.0%) with discrepant TP53 mutation and p53 IHC results, p53 IHC was repeated.
Results
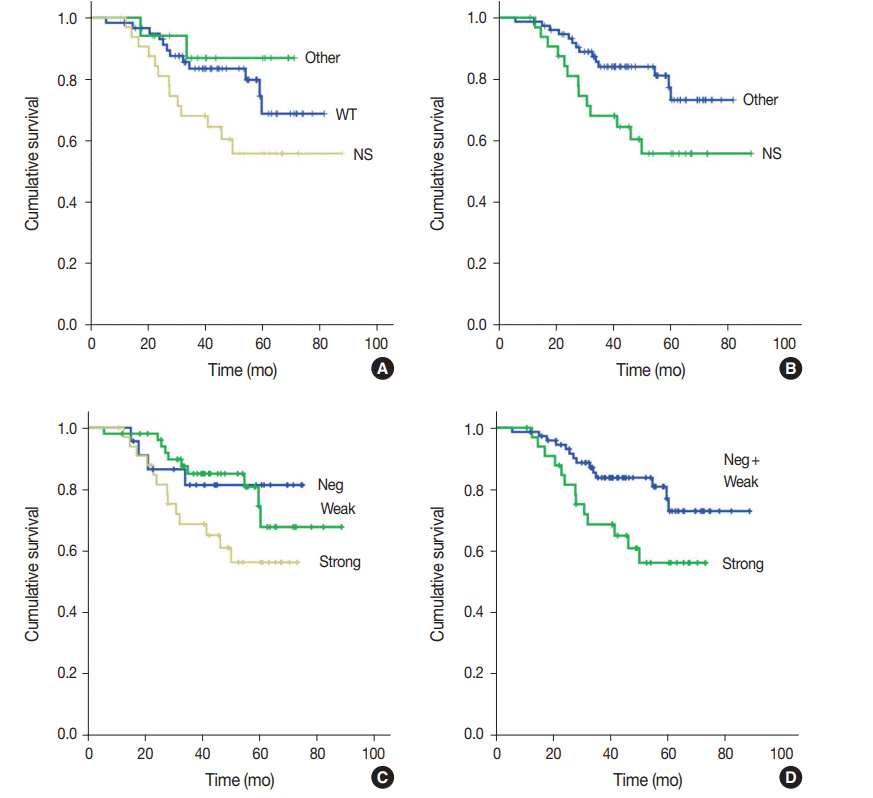
Strong expression of p53 was associated with TP53 missense mutations, negative expression with other types of mutations, and weak expression with wild-type TP53 (p<.001). The sensitivity for each category was 90.9%, 79.0%, and 80.9%, and the specificity was 95.4%, 88.1%, and 92.3%, respectively. The TNM stage at initial diagnosis exhibited a significant correlation with both TP53 mutation type (p=.004) and p53 expression status (p=.029). The Kaplan-Meier survival analysis for 109 stage II and III GC cases showed that patients with TP53 missense mutations had worse overall survival than those in the wild-type and other mutation groups (p=.028). Strong expression of p53 was also associated with worse overall survival in comparison to negative and weak expression (p=.035).
Conclusions
Results of IHC of the p53 protein may be used as a simple surrogate marker of TP53 mutations. However, negative expression of p53 and other types of mutations of TP53 should be carefully interpreted because of its lower sensitivity and different prognostic implications.
Figure
Cited by 1 articles
-
Clinicopathologic characterization of cervical metastasis from an unknown primary tumor: a multicenter study in Korea
Miseon Lee, Uiree Jo, Joon Seon Song, Youn Soo Lee, Chang Gok Woo, Dong-Hoon Kim, Jung Yeon Kim, Sun Och Yoon, Kyung-Ja Cho
J Pathol Transl Med. 2023;57(3):166-177. doi: 10.4132/jptm.2023.04.12.
Reference
-
References
1. Junttila MR, Evan GI. p53: a Jack of all trades but master of none. Nat Rev Cancer. 2009; 9:821–9.2. Bartkova J, Horejsi Z, Koed K, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005; 434:864–70.
Article3. Starzynska T, Bromley M, Ghosh A, Stern PL. Prognostic significance of p53 overexpression in gastric and colorectal carcinoma. Br J Cancer. 1992; 66:558–62.
Article4. Kakeji Y, Korenaga D, Tsujitani S, et al. Gastric cancer with p53 overexpression has high potential for metastasising to lymph nodes. Br J Cancer. 1993; 67:589–93.
Article5. Maehara Y, Tomoda M, Hasuda S, et al. Prognostic value of p53 protein expression for patients with gastric cancer: a multivariate analysis. Br J Cancer. 1999; 79:1255–61.6. Cancer Genome Atlas Research Network. Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014; 513:202–9.7. Cristescu R, Lee J, Nebozhyn M, et al. Molecular analysis of gastric cancer identifies subtypes associated with distinct clinical outcomes. Nat Med. 2015; 21:449–56.
Article8. Setia N, Agoston AT, Han HS, et al. A protein and mRNA expressionbased classification of gastric cancer. Mod Pathol. 2016; 29:772–84.
Article9. Ahn S, Lee SJ, Kim Y, et al. High-throughput Protein and mRNA Expression-based classification of gastric cancers can identify clinically distinct subtypes, concordant with recent molecular classifications. Am J Surg Pathol. 2017; 41:106–15.
Article10. Koh J, Lee KW, Nam SK, et al. Development and validation of an easy-to-implement, practical algorithm for the identification of molecular subtypes of gastric cancer: prognostic and therapeutic implications. Oncologist. 2019; 24:e1321–30.
Article11. Kobel M, Piskorz AM, Lee S, et al. Optimized p53 immunohistochemistry is an accurate predictor of TP53 mutation in ovarian carcinoma. J Pathol Clin Res. 2016; 2:247–58.12. Fukayama M, Rugge M, Washington MK. Tumours of the stomach. In : Lokuhetty D, White VA, Watanabe R, Cree IA, editors. WHO classification of tumours. Lyon: International Agency for Research on Cancer (IARC);2019. p. 85–103.13. Koh J, Nam SK, Roh H, et al. Somatic mutational profiles of stage II and III gastric cancer according to tumor microenvironment immune type. Genes Chromosomes Cancer. 2019; 58:12–22.
Article14. Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 2011; 17:10–2.
Article15. Cibulskis K, Lawrence MS, Carter SL, et al. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013; 31:213–9.
Article16. Cingolani P, Patel VM, Coon M, et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012; 3:35.
Article17. Liu X, Wu C, Li C, Boerwinkle E. dbNSFP v3.0: a one-stop database of functional predictions and annotations for human nonsynonymous and splice-site SNVs. Hum Mutat. 2016; 37:235–41.
Article18. Chakravarty D, Gao J, Phillips SM, et al. OncoKB: a precision oncology knowledge base. JCO Precis Oncol. 2017; 2017: 10.1200/PO.17.00011.
Article19. Landrum MJ, Lee JM, Benson M, et al. ClinVar: public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016; 44:D862–8.
Article20. Park Y, Koh J, Na HY, et al. PD-L1 testing in gastric cancer by the combined positive score of the 22C3 PharmDx and SP263 assay with clinically relevant cut-offs. Cancer Res Treat. 2020 Jan 10 [Epub]. https://doi.org/10.4143/crt.2019.718.
Article21. Song KY, Jung CK, Park WS, Park CH. Expression of the antiapoptosis gene survivin predicts poor prognosis of stage III gastric adenocarcinoma. Jpn J Clin Oncol. 2009; 39:290–6.
Article22. Mrena J, Wiksten JP, Kokkola A, Nordling S, Ristimaki A, Haglund C. COX-2 is associated with proliferation and apoptosis markers and serves as an independent prognostic factor in gastric cancer. Tumour Biol. 2010; 31:1–7.
Article23. Zha Y, Cun Y, Zhang Q, Li Y, Tan J. Prognostic value of expression of Kit67, p53, TopoIIa and GSTP1 for curatively resected advanced gastric cancer patients receiving adjuvant paclitaxel plus capecitabine chemotherapy. Hepatogastroenterology. 2012; 59:1327–32.24. Yildirim M, Kaya V, Demirpence O, Gunduz S, Bozcuk H. Prognostic significance of p53 in gastric cancer: a meta- analysis. Asian Pac J Cancer Prev. 2015; 16:327–32.25. Kobel M, Reuss A, du Bois A, et al. The biological and clinical value of p53 expression in pelvic high-grade serous carcinomas. J Pathol. 2010; 222:191–8.26. Shin YJ, Kim Y, Wen X, et al. Prognostic implications and interaction of L1 methylation and p53 expression statuses in advanced gastric cancer. Clin Epigenetics. 2019; 11:77.
Article27. Nenutil R, Smardova J, Pavlova S, et al. Discriminating functional and non-functional p53 in human tumours by p53 and MDM2 immunohistochemistry. J Pathol. 2005; 207:251–9.
Article28. Watanabe G, Ishida T, Furuta A, et al. Combined immunohistochemistry of PLK1, p21, and p53 for predicting TP53 status: an independent prognostic factor of breast cancer. Am J Surg Pathol. 2015; 39:1026–34.29. Tahara T, Shibata T, Okamoto Y, et al. Mutation spectrum of TP53 gene predicts clinicopathological features and survival of gastric cancer. Oncotarget. 2016; 7:42252–60.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Prognostic Significance of p53 Protein and PCNA in Advanced Gastric Carcinoma
- p53 Gene Mutation in Gastric Cancer Tissue
- Somatic Mutations of TP53 Identified by Targeted Next-Generation Sequencing Are Poor Prognostic Factors for Primary Operable Breast Cancer: A Single-Center Study
- The Clinical Significance of Mutation in the p53, DCC and nm23 Genes in Patients with Gastric Carcinoma
- p53 Mutation in Gastric Carcinoma Detected by PCR - SSCP and Direct - Sequencing