J Vet Sci.
2020 Jan;21(1):e4. 10.4142/jvs.2020.21.e4.
Rapid detection of deformed wing virus in honeybee using ultra-rapid qPCR and a DNA-chip
- Affiliations
-
- 1Department of Life Science, College of Fusion Science, Kyonggi University, Suwon 16227, Korea. ryheekim@gmail.com
- KMID: 2469019
- DOI: http://doi.org/10.4142/jvs.2020.21.e4
Abstract
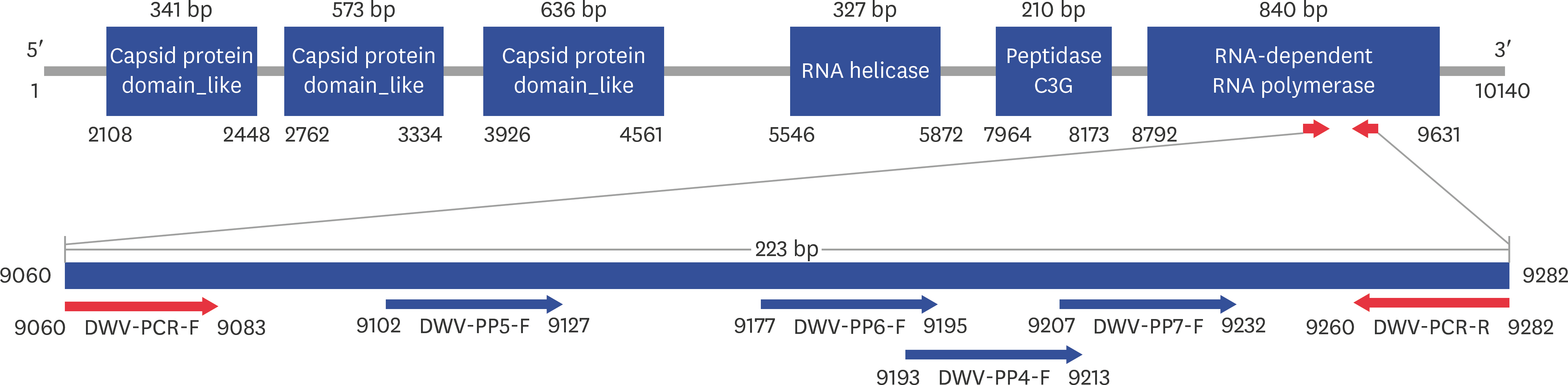
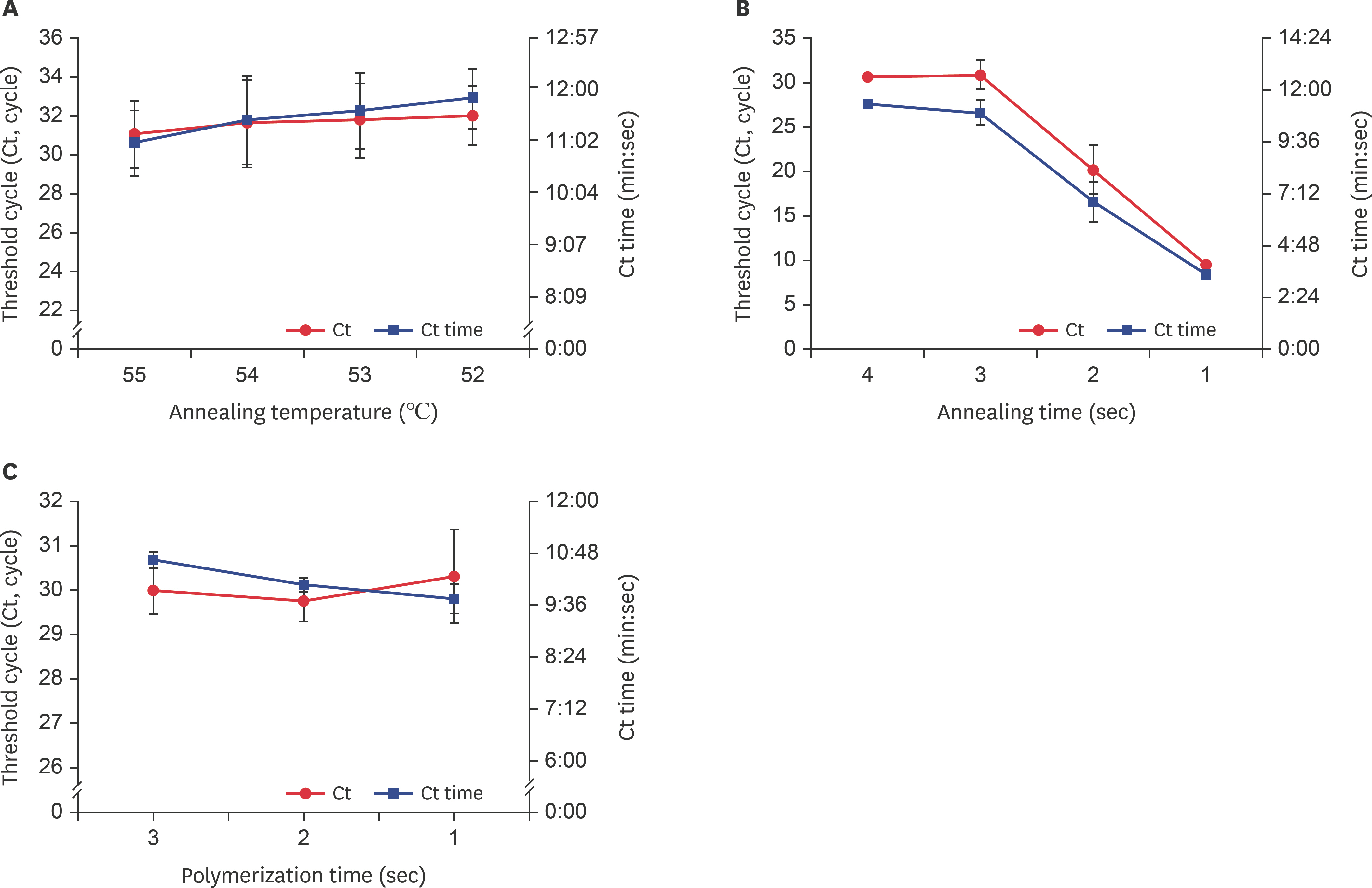
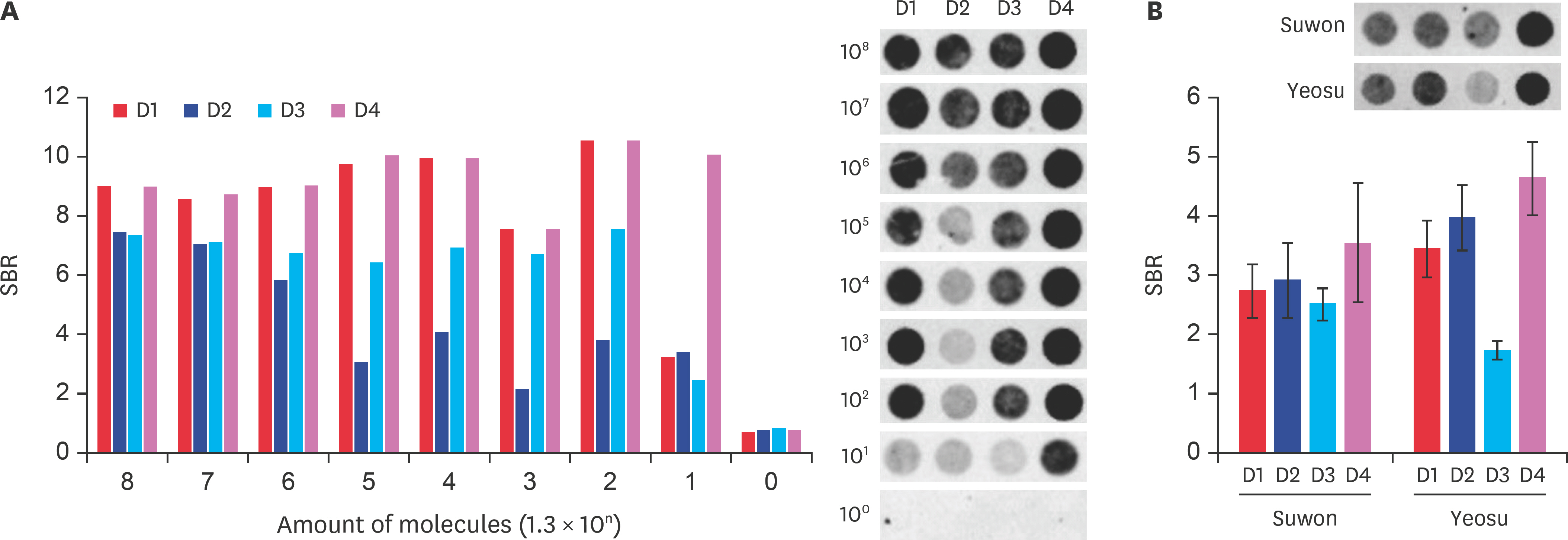
- Fast and accurate detection of viral RNA pathogens is important in apiculture. A polymerase chain reaction (PCR)-based detection method has been developed, which is simple, specific, and sensitive. In this study, we rapidly (in 1 min) synthesized cDNA from the RNA of deformed wing virus (DWV)-infected bees (Apis mellifera), and then, within 10 min, amplified the target cDNA by ultra-rapid qPCR. The PCR products were hybridized to a DNA-chip for confirmation of target gene specificity. The results of this study suggest that our method might be a useful tool for detecting DWV, as well as for the diagnosis of RNA virus-mediated diseases on-site.
MeSH Terms
Figure
Reference
-
References
1. McMenamin AJ, Genersch E. Honey bee colony losses and associated viruses. Curr Opin Insect Sci. 2015; 8:121–129.
Article2. Genersch E, Yue C, Fries I, de Miranda JR. Detection of Deformed wing virus, a honey bee viral pathogen, in bumble bees (Bombus terrestris and Bombus pascuorum) with wing deformities. J Invertebr Pathol. 2006; 91:61–63.3. Tehel A, Brown MJ, Paxton RJ. Impact of managed honey bee viruses on wild bees. Curr Opin Virol. 2016; 19:16–22.
Article4. Reddy KE, Noh JH, Yoo MS, Kim YH, Kim NH, Doan HT, Ramya M, Jung SC, Van Quyen D, Kang SW. Molecular characterization and phylogenetic analysis of deformed wing viruses isolated from South Korea. Vet Microbiol. 2013; 167:272–279.
Article5. Tentcheva D, Gauthier L, Jouve S, Canabady-Rochelle L, Dainat B, Cousserans F, Colina ME, Ballb BV, Bergoin M. Polymerase chain reaction detection of deformed wing virus (DWV) in Apis mellifera and Varroa destructor. Apidologie (Celle). 2004; 35:431–439.6. Chen YP, Higgins JA, Feldlaufer MF. Quantitative real-time reverse transcription-PCR analysis of deformed wing virus infection in the honeybee (Apis mellifera L.). Appl Environ Microbiol. 2005; 71:436–441.7. Yoo MS, Thi KC, Van Nguyen P, Han SH, Kwon SH, Yoon BS. Rapid detection of sacbrood virus in honeybee using ultra-rapid real-time polymerase chain reaction. J Virol Methods. 2012; 179:195–200.
Article8. Jamnikar Cigleneč ki U, Toplak I. Development of a real-time RT-PCR assay with TaqMan probe for specific detection of acute bee paralysis virus. J Virol Methods. 2012; 184:63–68.
Article9. Haddad NJ, Noureddine A, Al-Shagour B, Loucif-Ayad W, El-Niweiri MA, Anaswah E, Hammour WA, El-Obeid D, Imad A, Shebl MA, Almaleky AS, Nasher A, Walid N, Bergigui MF, Yañez O, de Miranda JR. Distribution and variability of deformed wing virus of honeybees (Apis mellifera) in the Middle East and North Africa. Insect Sci. 2017; 24:103–113.10. Kukielka D, Esperón F, Higes M, Sánchez-Vizcaíno JM. A sensitive one-step real-time RT-PCR method for detection of deformed wing virus and black queen cell virus in honeybee Apis mellifera. J Virol Methods. 2008; 147:275–281.11. Lim HY. Development of novel rapid detection method for deformed wing virus (DWV) using ultra-fast high-performance PCR (UF-HP PCR). J Apic. 2013; 28:237–244.12. Ma M, Ma C, Li M, Wang S, Yang S, Wang S. Loop-mediated isothermal amplification for rapid detection of Chinese sacbrood virus. J Virol Methods. 2011; 176:115–119.
Article13. Yoo MS, Noh JH, Yoon BS, Reddy KE, Kweon CH, Jung SC, Kang SW. Reverse transcription loop-mediated isothermal amplification for sensitive and rapid detection of Korean sacbrood virus. J Virol Methods. 2012; 186:147–151.
Article14. Han SH, Lee DB, Lee DW, Kim EH, Yoon BS. Ultra-rapid real-time PCR for the detection of Paenibacillus larvae, the causative agent of American Foulbrood (AFB). J Invertebr Pathol. 2008; 99:8–13.15. Lee DW, Kim EH, Yoo MS, Han SH. Ultra-rapid real-time PCR for the detection of human immunodeficiency virus (HIV). Korean J Microbiol. 2007; 43:91–99.16. Lim SJ, Kim JM, Lee CW, Yoon BS. Development of ultra-rapid multiplex PCR detection against 6 major pathogens in honeybee. J Apic. 2017; 32:27–39.
Article17. Lung O, Fisher M, Beeston A, Hughes KB, Clavijo A, Goolia M, Pasick J, Mauro W, Deregt D. Multiplex RT-PCR detection and microarray typing of vesicular disease viruses. J Virol Methods. 2011; 175:236–245.
Article18. Choe SE, Nguyen LT, Noh JH, Koh HB, Jean YH, Kweon CH, Kang SW. Prevalence and distribution of six bee viruses in Korean Apis cerana populations. J Invertebr Pathol. 2012; 109:330–333.19. Tantillo G, Bottaro M, Di Pinto A, Martella V, Di Pinto P, Terio V. Virus infections of honeybees Apis Mellifera. Ital J Food Saf. 2015; 4:5364.20. Cai HY, Caswell JL, Prescott JF. Nonculture molecular techniques for diagnosis of bacterial disease in animals: a diagnostic laboratory perspective. Vet Pathol. 2014; 51:341–350.21. Wang JH, Lee DB, Ku SJ, Peak MC, Min SH, Lim SJ, Lee CW, Yoon BS. Development of a detection method against 11 major pathogens of honey bee using amplification of multiplex PCR and specific DNA-chip. J Apic. 2016; 31:133–146.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Rapidly quantitative detection of Nosema ceranae in honeybees using ultra-rapid real-time quantitative PCR
- A Case of Ultra-rapid Cycling Bipolar Disorder Recovered by Lamotrigine Combination
- Detection of rpoB Gene Mutation in Rifampin-Resistant M. Tuberculosis by Oligonucleotide Chip
- Prevalence of Nosema and Virus in Honey Bee (Apis mellifera L.) Colonies on Flowering Period of Acacia in Korea
- Comparison of Rapid Antigen Test and Real-Time Reverse Transcription PCR for the Detection of Influenza B Virus