J Bacteriol Virol.
2019 Dec;49(4):212-220. 10.4167/jbv.2019.49.4.212.
Identification of Candida Species Using 26S Ribosomal RNA Gene Sequencing in Patients with Periodontitis
- Affiliations
-
- 1Department of Microbiology, College of Dentistry, University of Babylon, Babil 51001, Iraq.
- 2Department of Animal Production, College of Agriculture, Al-Qasim Green University, Babil, 51001, Iraq. mohammed79@agre.uoqasim.edu.iq, baquralhilly_79@yahoo.com
- KMID: 2468027
- DOI: http://doi.org/10.4167/jbv.2019.49.4.212
Abstract
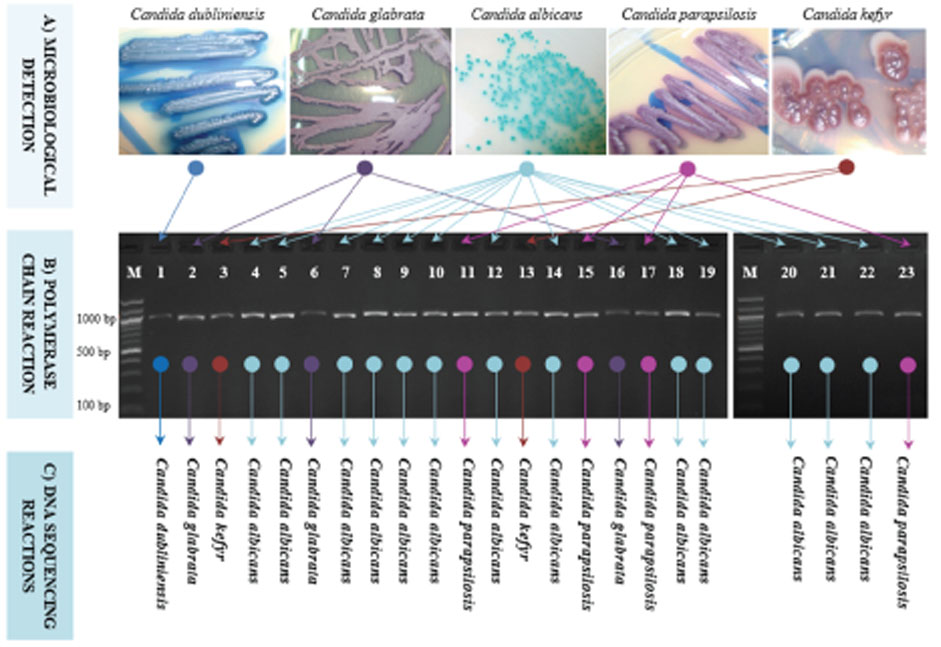
- The infection with Candida spp. for oral cavity is being increasingly reported. However, its variations have not yet been specifically described in periodontitis. The present study was conducted to use an uniplex 26S rRNA-based amplicons to detect and discriminate Candida using only one pair of ribosomal primers. A total of 50 patients with chronic periodontitis was involved in the study. Pure Candida colonies were isolated from 23 patients and genomic DNA was extracted, and PCR was conducted. Direct DNA sequencing followed by comprehensive phylogenetic analyses were performed to confirm the identity of Candida colonies. Results indicated that the ration of Candida-infected patients was 46%, with a high prevalence of C. albicans, followed by remarkably lower ratios of C. parapsilosis, C. glabrata, C. kefyr, and C. dubliniensis respectively. Phylogenetic analyses indicated obvious discrimination amongst the analyzed Candida species as each observed species occupied a distinctive phylogenetic position. The current results reported a simple, efficient, and low-cost detection of five species of Candida without the need for other costly techniques of molecular screening. The current findings may help dentists to easily take a snapshot of the patterns of Candida infection in periodontitis cases to assess the nature and grade of infection.
MeSH Terms
Figure
Reference
-
1. Turner SA, Butler G. The Candida pathogenic species complex. Cold Spring Harb Perspect Med. 2014; 4:a019778.2. Araújo D, Henriques M, Silva S. Portrait of Candida species biofilm regulatory network genes. Trends Microbiol. 2017; 25:62–75.3. Aggarwal N, Bhateja S, Arora G, Yasmin T. Candidiasis- The most common fungal infection of oral cavity. Biomed J Sci Tech Res. 2018; 8:6487–6491.4. Thiyahuddin NM, Lamping E, Rich AM, Cannon RD. Yeast Species in the Oral Cavities of Older People: A Comparison between People Living in Their Own Homes and Those in Rest Homes. J Fungi (Basel). 2019; 5:E30.
Article5. Zahir RA, Himratul-Aznita WH. Distribution of Candida in the oral cavity and its differentiation based on the internally transcribed spacer (ITS) regions of rDNA. Yeast. 2013; 30:13–23.
Article6. Garcia-Cuesta C, Sarrion-Pérez MG, Bagán JV. Current treatment of oral candidiasis: A literature review. J Clin Exp Dent. 2014; 6:e576–e582.
Article7. Niimi M, Cannon RD, Monk BC. Candida albicans pathogenicity: a proteomic perspective. Electrophoresis. 1999; 20:2299–2308.
Article8. Whaley SG, Berkow EL, Rybak JM, Nishimoto AT, Barker KS, Rogers PD. Azole antifungal resistance in Candida albicans and emerging non-albicans Candida species. Front Microbiol. 2017; 7:2173.9. Akram A, Maley M, Gosbell I, Nguyen T, Chavada R. Utility of 16S rRNA PCR performed on clinical specimens in patient management. Int J Infect Dis. 2017; 57:144–149.
Article10. Al-Shuhaib MBS, Al-Kaaby HN, Alwan SL. A highly efficient electrophoretic method for discrimination between two Neoscytalidium species using a specific fungal internal transcribed spacer (ITS) fragment. Folia Microbiol (Praha). 2019; 64:161–170.
Article11. Al-Dabbagh NN, Hashim HO, Al-Shuhaib MBS. A highly efficient computational discrimination among Streptococcal species of periodontitis patients using 16S rRNA amplicons. Korean J Microbiol. 2019; 55:1–8.12. Diezmann S, Cox CJ, Schönian G, Vilgalys RJ, Mitchell TG. Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J Clin Microbiol. 2004; 42:5624–5635.
Article13. Venkatesan G, Uppoor A, Naik D, Kadkampally D, Maddi A. Oral Candida Carriage and Morphotype Differentiation in Chronic Periodontitis Patients with and without Diabetes in the Indian Sub-Continent. Dent J (Basel). 2015; 3:123–131.
Article14. Gomes CC, Guimarães LS, Pinto LCC, Camargo GADCG, Valente MIB, Sarquis MIM. Investigations of the prevalence and virulence of Candida albicans in periodontal and endodontic lesions in diabetic and normoglycemic patients. J Appl Oral Sci. 2017; 25:274–281.
Article15. Boonsilp S, Thaipadungpanit J, Amornchai P, Wuthiekanun V, Chierakul W, Limmathurotsakul D, Day NP, Peacock SJ. Molecular detection and speciation of pathogenic Leptospira spp. in blood from patients with culture-negative leptospirosis. BMC Infect Dis. 2011; 11:338.16. Canabarro A, Valle C, Farias MR, Santos FB, Lazera M, Wanke B. Association of subgingival colonization of Candida albicans and other yeasts with severity of chronic periodontitis. J Periodontal Res. 2013; 48:428–432.
Article17. Ito CY, de Paiva Martins CA, Loberto JC, dos Santos SS, Jorge AO. In vitro antifungal susceptibility of Candida spp. isolates from patients with chronic periodontitis and from control patients. Braz Oral Res. 2004; 18:80–84.
Article18. Loberto JCS, Martins CAP, Santos SSF, Cortelli JR, Jorge AOC. Staphylococcus spp. in the oral cavity and periodontal pockets of chronic periodontitis patients. Braz J Microbiol. 2004; 35:64–68.19. Kim D, Shin WS, Lee KH, Kim K, Park JY, Koh CM. Rapid differentiation of Candida albicans from other Candida species using its unique germ tube formation at 39°C. Yeast. 2002; 19:957–962.
Article20. Al-Shuhaib MBS, Albakri AH, Alwan SH, Almandil NB, AbdulAzeez S, Borgio FJ. Optimal PCR primers for rapid and accurate detection of Aspergillus flavus isolates. Microb Pathog. 2018; 116:351–355.
Article21. Nadeem SG, Hakim ST, Kazmi SU. Use of CHROMagar Candida for the presumptive identification of Candida species directly from clinical specimens in resource-limited settings. Libyan J Med. 2010; 5:2144.
Article22. Pravin Charles MV, Kali A, Joseph NM. Performance of chromogenic media for Candida in rapid presumptive identification of Candida species from clinical materials. Pharmacognosy Res. 2015; 7:Suppl 1. S69–S73.23. Sahand IH, Moragues MD, Eraso E, Villar-Vidal M, Quindós G, Pontón J. Supplementation of CHROMagar Candida medium with Pal's medium for rapid identification of Candida dubliniensis. J Clin Microbiol. 2005; 43:5768–5770.
Article24. Dufresne SF, Marr KA, Sydnor E, Staab JF, Karp JE, Lu K, et al. Epidemiology of Candida kefyr in patients with hematologic malignancies. J Clin Microbiol. 2014; 52:1830–1837.
Article25. Singh A, Verma R, Murari A, Agrawal A. Oral candidiasis: An overview. J Oral Maxillofac Pathol. 2014; 18:Suppl 1. S81–S85.
Article26. Zunt SL. Oral candidiasis: diagnosis and treatment. The Journal of Practical Hygiene. 2000; 9:31–36.27. Zuza-Alves DL, Silva-Rocha WP, Chaves GM. An update on Candida tropicalis based on basic and clinical approaches. Front Microbiol. 2017; 8:1927.28. Zanni PCMD, Bonfim-Mendonça PS, Negri M, Nakamura SS, Donatti L, Svidzinski TIE, et al. Virulence factors and genetic variability of vaginal Candida albicans isolates from HIV-infected women in the post-highly active antiretroviral era. Rev Inst Med Trop Sao Paulo. 2017; 59:e44.
Article29. Hewitt SK, Foster DS, Dyer PS, Avery SV. Phenotypic heterogeneity in fungi: Importance and methodology. Fungal Biol Rev. 2016; 30:176–184.
Article30. Dhanasekaran D, Vinothini K, Latha S, Thajuddin N, Panneerselvam A. Human dental biofilm: Screening, characterization, in vitro biofilm formation and antifungal resistance of Candida spp. Saudi J Dent Sci. 2014; 5:55–70.
Article31. Arastehfar A, Fang W, Pan W, Lackner M, Liao W, Badiee P, et al. YEAST PANEL multiplex PCR for identification of clini cally important yeast species: stepwise diagnostic strategy, useful for developing countries. Diagn Microbiol Infect Dis. 2019; 93:112–119.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Laboratory Identification of Leptotrichia Species Isolated From Bacteremia Patients at a Single Institution
- Two New Species of Cryptococcus sp. and Candida sp. from Wild Flowers in Korea
- Three Cases of Moraxella osloensis Meningitis: A Difficult Experience in Species Identification and Determination of Clinical Significance
- The Application of 26S rDNA PCR-RFLP in the Identification and Classification of Malassezia Yeast
- A riboprinting scheme for identification of unknown Acanthamoeba isolates at species level