Healthc Inform Res.
2019 Apr;25(2):131-138. 10.4258/hir.2019.25.2.131.
Development of Artificial Intelligence to Support Needle Electromyography Diagnostic Analysis
- Affiliations
-
- 1Department of Biomedical Engineering, Chungnam National University Graduade School, Daejeon, Korea.
- 2Department of Rehabilitation Medicine, Chungnam National University Hospital, Daejeon, Korea. 102onez@cnuh.co.kr
- 3Department of Rehabilitation Medicine, Chungnam National University College of Medicine, Daejeon, Korea.
- 4Department of Biomedical Engineering, Chungnam National University College of Medicine, Daejeon, Korea.
- KMID: 2445010
- DOI: http://doi.org/10.4258/hir.2019.25.2.131
Abstract
OBJECTIVES
This study proposes a method for classifying three types of resting membrane potential signals obtained as images through diagnostic needle electromyography (EMG) using TensorFlow-Slim and Python to implement an artificial-intelligence-based image recognition scheme.
METHODS
Waveform images of an abnormal resting membrane potential generated by diagnostic needle EMG were classified into three types"”positive sharp waves (PSW), fibrillations (Fibs), and Others"”using the TensorFlow-Slim image classification model library. A total of 4,015 raw waveform data instances were reviewed, with 8,576 waveform images subsequently collected for training. Images were learned repeatedly through a convolutional neural network. Each selected waveform image was classified into one of the aforementioned categories according to the learned results.
RESULTS
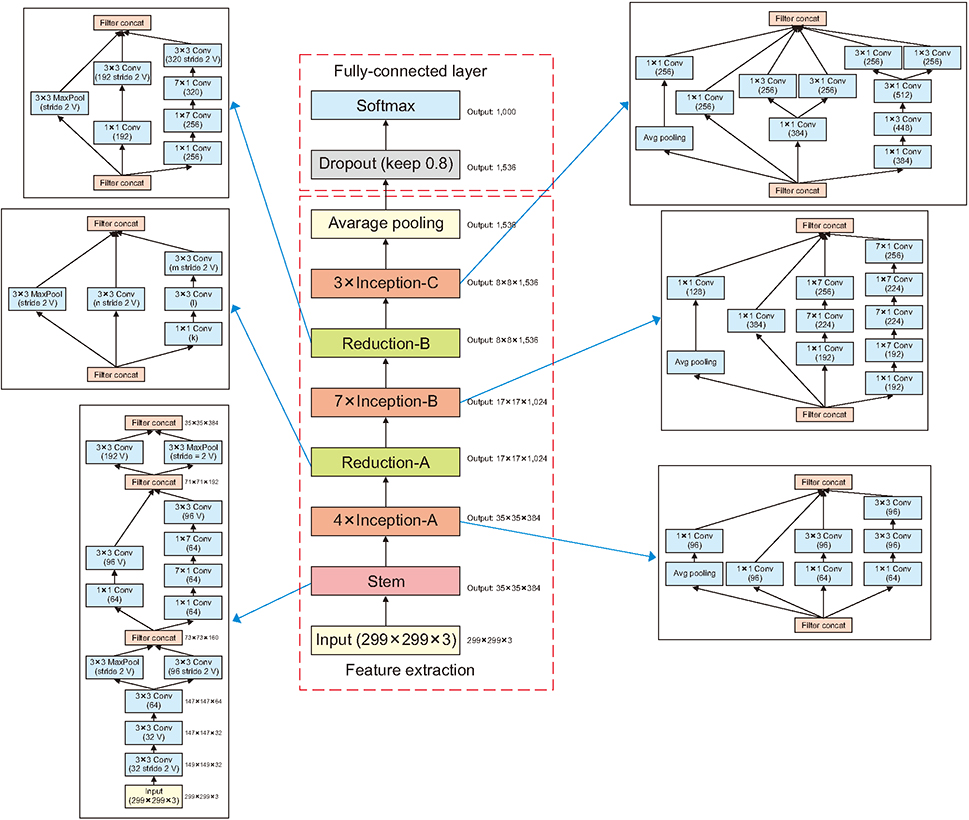
The classification model, Inception v4, was used to divide waveform images into three categories (accuracy = 93.8%, precision = 99.5%, recall = 90.8%). This was done by applying the pretrained Inception v4 model to a fine-tuning method. The image recognition model was created for training using various types of image-based medical data.
CONCLUSIONS
The TensorFlow-Slim library can be used to train and recognize image data, such as EMG waveforms, through simple coding rather than by applying TensorFlow. It is expected that a convolutional neural network can be applied to image data such as the waveforms of electrophysiological signals in a body based on this study.
Keyword
MeSH Terms
Figure
Reference
-
1. Lindstrom H, Ashworth NL. The usefulness of electrodiagnostic studies in the diagnosis and management of neuromuscular disorders. Muscle Nerve. 2018; 58(2):191–196.
Article2. Noorbakhsh-Sabet N, Zand R, Zhang Y, Abedi V. Artificial intelligence transforms the future of health care. Am J Med. 2019; 01. 31. [Epub]. DOI: 10.1016/j.amjmed.2019.01.017.
Article3. NVIDIA CUDA Toolkit [Internet]. Santa Clara (CA): NVIDIA Corp.;c2019. cited as 2019 Jan 15. Available from: https://developer.nvidia.com/cuda-toolkit.4. TensorFlow.org. GPU support [Internet]. [place unknown]: TensorFlow.org;c2019. cited as 2019 Jan 15. Available from: https://www.tensorflow.org/install/gpu.5. NVIDIA cuDNN [Internet]. Santa Clara (CA): NVIDIA Corp.;c2019. cited as 2019 Jan 15. Available from: https://developer.nvidia.com/cudnn.6. TensorFlow.org. Install TensorFlow with pip [Internet]. [place unknown]: TensorFlow.org;2015. cited at 2019 Apr 15. Available from: https://www.tensorflow.org/install/pip?hl=ko.7. Silberman N, Guadarrama S. TensorFlow-Slim image classification model library [Internet]. [place unknown]: GitHub Inc.;2016. cited at 2019 Apr 15. Available from: https://github.com/tensorflow/models/tree/master/research/slim.8. GitHub Inc. TensorFlow models [Internet]. [place unknown]: GitHub Inc.;c2019. cited at 2019 Apr 15. Available from: https://github.com/tensorflow/models.9. Szegedy C, Ioffe S, Vanhoucke V, Alemi AA. Inceptionv4, Inception-ResNet and the impact of residual connections on learning. In : Proceedings of the 31st AAAI Conference on Artificial Intelligence; 2017 Feb 4-9; San Francisco, CA. p. 4278–4284.10. Pham TC, Luong CM, Visani M, Hoang VD. Deep CNN and data augmentation for skin lesion classification. In : Nguyen N, Hoang D, Hong TP, Pham H, Trawinski B, editors. Intelligent information and database systems. Cham, Switzerland: Springer;2018. p. 573–582.11. GitHub Inc. Inception[3] Inception v4, Inception ResNet and the Impact of Residual Connections on Learning(2016) [Internet]. [place unknown]: GitHub Inc.;2018. cited at 2019 Apr 15. Available from: https://github.com/hwkim94/hwkim94.github.io/wiki.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Role of artificial intelligence in diagnosing Barrett’s esophagus-related neoplasia
- The Role of medical doctor in the era of artificial intelligence
- Application of artificial intelligence for diagnosis of early gastric cancer based on magnifying endoscopy with narrow-band imaging
- The future of artificial intelligence for physicians
- Artificial Intelligence in Pathology