Blood Res.
2018 Dec;53(4):320-324. 10.5045/br.2018.53.4.320.
Altered expression of MALAT1 lncRNA in chronic lymphocytic leukemia patients, correlation with cytogenetic findings
- Affiliations
-
- 1Department of Hematology, Faculty of Medical Sciences, Tarbiat Modarres University, Tehran, Iran. kavianis@modares.a.ir
- 2Hematology-Oncology and Stem Cell Transplantation Research Center, Tehran, Iran. m-yaghmaie@sina.tums.ac.ir
- 3Cell Therapy and Hematopoietic Stem Cell Transplantation Research Center, Tehran, Iran.
- 4Hematologic Malignancies Research Center, Tehran University of Medical Sciences, Tehran, Iran.
- 5Comprehensive Cancer Research Center, Mazandaran University of Medical Science, Sari, Iran.
- KMID: 2429307
- DOI: http://doi.org/10.5045/br.2018.53.4.320
Abstract
- BACKGROUND
Recent studies have devoted much attention to non-protein-coding transcripts in relation to a wide range of malignancies. MALAT1, a long non-coding RNA, has been reported to be associated with cancer progression and prognosis. Thus, we here determined MALAT1 gene expression in chronic lymphocytic leukemia (CLL), a genetically heterogeneous disease, and explored its possible relationships with cytogenetic abnormalities.
METHODS
MALAT1 expression level was evaluated using real-time quantitative reverse transcription polymerase chain reaction (qRT-PCR) on blood mononuclear cells from 30 non-treated CLL patients and 30 matched healthy controls. Cytogenetic abnormalities were determined in patients by fluorescence in situ hybridization (FISH).
RESULTS
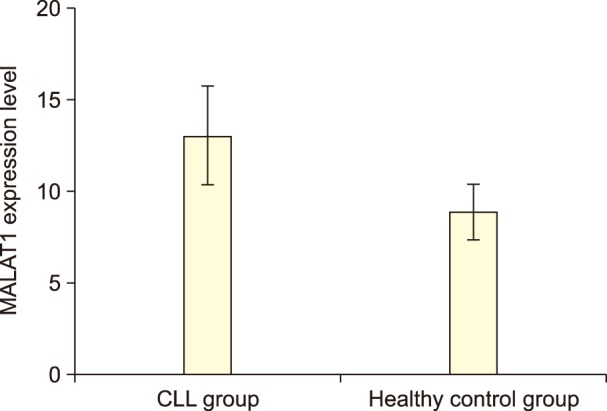
MALAT1 expression level was up-regulated in the CLL group compared to healthy controls (P=0.008). Del13q14, followed by Del11q22, were the most prevalent cytogenetic abnormalities. We found no association between the FISH results and MALAT1 expression in patients.
CONCLUSION
Altered expression of MALAT1 is associated with CLL development. Further investigations are required to assess the relationship between this long non-coding RNA and CLL patient survival and prognosis.
Keyword
MeSH Terms
Figure
Reference
-
1. Herishanu Y, Katz BZ, Lipsky A, Wiestner A. Biology of chronic lymphocytic leukemia in different microenvironments: clinical and therapeutic implications. Hematol Oncol Clin North Am. 2013; 27:173–206. PMID: 23561469.2. Bauer K, Rancea M, Roloff V, et al. Rituximab, ofatumumab and other monoclonal anti-CD20 antibodies for chronic lymphocytic leukaemia. Cochrane Database Syst Rev. 2012; 11:CD008079. PMID: 23152253.
Article3. Charbotel B, Fervers B, Droz JP. Occupational exposures in rare cancers: A critical review of the literature. Crit Rev Oncol Hematol. 2014; 90:99–134. PMID: 24387944.
Article4. Parker TL, Strout MP. Chronic lymphocytic leukemia: prognostic factors and impact on treatment. Discov Med. 2011; 11:115–123. PMID: 21356166.5. Inamura K. Major tumor suppressor and oncogenic non-coding RNAs: clinical relevance in lung cancer. Cells. 2017; 6:E12. PMID: 28486418.
Article6. Taheri M, Habibi M, Noroozi R, et al. HOTAIR genetic variants are associated with prostate cancer and benign prostate hyperplasia in an Iranian population. Gene. 2017; 613:20–24. PMID: 28259691.
Article7. Tripathi V, Shen Z, Chakraborty A, et al. Long noncoding RNA MALAT1 controls cell cycle progression by regulating the expression of oncogenic transcription factor B-MYB. PLoS Genet. 2013; 9:e1003368. PMID: 23555285.
Article8. Ahmadi J, Kaviani Gebelli S, Atashi A. Evaluation of MALAT1 gene expression in AML and ALL cell lines. Faslnamahi Kumish. 2015; 17:179–186.9. Cho SF, Chang YC, Chang CS, et al. MALAT1 long non-coding RNA is overexpressed in multiple myeloma and may serve as a marker to predict disease progression. BMC Cancer. 2014; 14:809. PMID: 25369863.
Article10. Janbabaei G, Hedayatizadeh-Omran A, Alizadeh-Navaei R, et al. An epidemiological study of patients with breast cancer in Northern Iran, between 2006 and 2015. WCRJ. 2016; 3:e803.11. Dykes IM, Emanueli C. Transcriptional and post-transcriptional gene regulation by long non-coding RNA. Genomics Proteomics Bioinformatics. 2017; 15:177–186. PMID: 28529100.
Article12. Li J, Xuan Z, Liu C. Long non-coding RNAs and complex human diseases. Int J Mol Sci. 2013; 14:18790–18808. PMID: 24036441.
Article13. Eftekharian MM, Ghafouri-Fard S, Soudyab M, et al. Expression analysis of long non-coding RNAs in the blood of multiple sclerosis patients. J Mol Neurosci. 2017; 63:333–341. PMID: 28967047.
Article14. Rahimi H, Sadeghian MH, Keramati MR, et al. Cytogenetic abnormalities with interphase FISH method and clinical manifestation in chronic lymphocytic leukemia patients in North-East of Iran. Int J Hematol Oncol Stem Cell Res. 2017; 11:217–224. PMID: 28989588.15. Balatti V, Pekarky Y, Rizzotto L, Croce CM. miR deregulation in CLL. Adv Exp Med Biol. 2013; 792:309–325. PMID: 24014303.
Article16. Sadighi S, Jahanzad I, Ali Mohagheghi M, et al. Somatic mutation in immunoglobulin gene variable region in patients with chronic lymphoid leukemia and its influence on disease prognosis. Middle East J Rehabil Health Stud. 2016; 2:e35848.
Article17. Ronchetti D, Manzoni M, Agnelli L, et al. lncRNA profiling in early-stage chronic lymphocytic leukemia identifies transcriptional fingerprints with relevance in clinical outcome. Blood Cancer J. 2016; 6:e468. PMID: 27611921.
Article18. Amodio N, Stamato MA, Juli G, et al. Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity. Leukemia. 2018; 32:1948–1957. PMID: 29487387.
Article19. Lamothe B, Cervantes-Gomez F, Sivina M, Wierda WG, Keating MJ, Gandhi V. Proteasome inhibitor carfilzomib complements ibrutinib’s action in chronic lymphocytic leukemia. Blood. 2015; 125:407–410. PMID: 25573971.
Article20. Kuhn DJ, Chen Q, Voorhees PM, et al. Potent activity of carfilzomib, a novel, irreversible inhibitor of the ubiquitin-proteasome pathway, against preclinical models of multiple myeloma. Blood. 2007; 110:3281–3290. PMID: 17591945.
Article22. Kay NE, Suen R, Ranheim E, Peterson LC. Confirmation of Rb gene defects in B-CLL clones and evidence for variable predominance of the Rb defective cells within the CLL clone. Br J Haematol. 1993; 84:257–264. PMID: 8398827.
Article23. Huang JL, Liu W, Tian LH, et al. Upregulation of long non-coding RNA MALAT-1 confers poor prognosis and influences cell proliferation and apoptosis in acute monocytic leukemia. Oncol Rep. 2017; 38:1353–1362. PMID: 28713913.
Article24. Isin M, Ozgur E, Cetin G, et al. Investigation of circulating lncRNAs in B-cell neoplasms. Clin Chim Acta. 2014; 431:255–259. PMID: 24583225.
Article25. Wang X, Sehgal L, Jain N, Khashab T, Mathur R, Samaniego F. LncRNA MALAT1 promotes development of mantle cell lymphoma by associating with EZH2. J Transl Med. 2016; 14:346. PMID: 27998273.
Article26. Li LJ, Chai Y, Guo XJ, Chu SL, Zhang LS. The effects of the long non-coding RNA MALAT-1 regulated autophagy-related signaling pathway on chemotherapy resistance in diffuse large B-cell lymphoma. Biomed Pharmacother. 2017; 89:939–948. PMID: 28292022.
Article27. Kim SH, Kim SH, Yang WI, Kim SJ, Yoon SO. Association of the long non-coding RNA MALAT1 with the polycomb repressive complex pathway in T and NK cell lymphoma. Oncotarget. 2017; 8:31305–31317. PMID: 28412742.
Article28. Kokhaee P. B-cell chronic lymphocyte leukemia (B-CLL). Faslnamahi Kumish. 2007; 9:1–12.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Silencing of Long Non-Coding RNA MALAT1 Promotes Apoptosis of Glioma Cells
- Long Noncoding RNA MALAT1 Regulates Hepatocellular Carcinoma Growth Under Hypoxia via Sponging MicroRNA-200a
- First Case of Transformation of Immunoglobulin Heavy Chain Variable-Mutated Chronic Lymphocytic Leukemia Into Chronic Myeloid Leukemia
- A Case of Leukemia Cutis Associated with B-cell Chronic Lymphocytic Leukemia
- Leukemic Macrocheilitis Associated with Chronic Lymphocytic Leukemia