Transl Clin Pharmacol.
2017 Dec;25(4):166-172. 10.12793/tcp.2017.25.4.166.
Screening study for genetic polymorphisms affecting pharmacokinetics of talniflumate
- Affiliations
-
- 1Department of Clinical Pharmacology and Therapeutics, College of Medicine, Kyung Hee University, Seoul 02447, Korea. ysvin@khu.ac.kr
- 2Department of BioMedical Science, College of Life Science, CHA University, SeongNam 13488, Republic of Korea. kbkwack@cha.ac.kr
- KMID: 2406997
- DOI: http://doi.org/10.12793/tcp.2017.25.4.166
Abstract
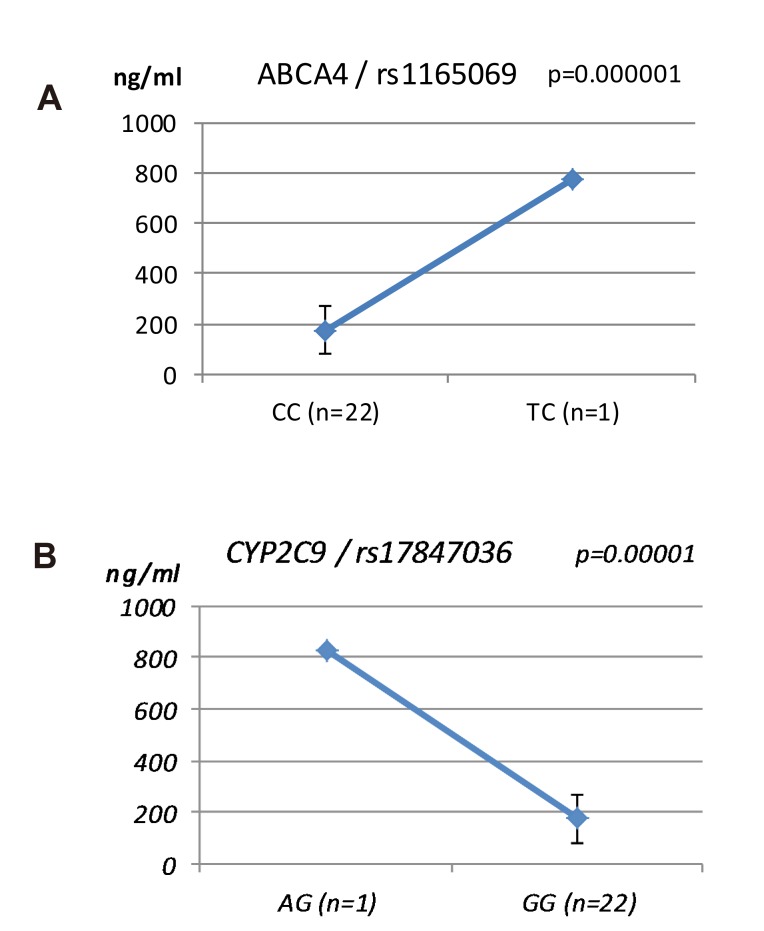
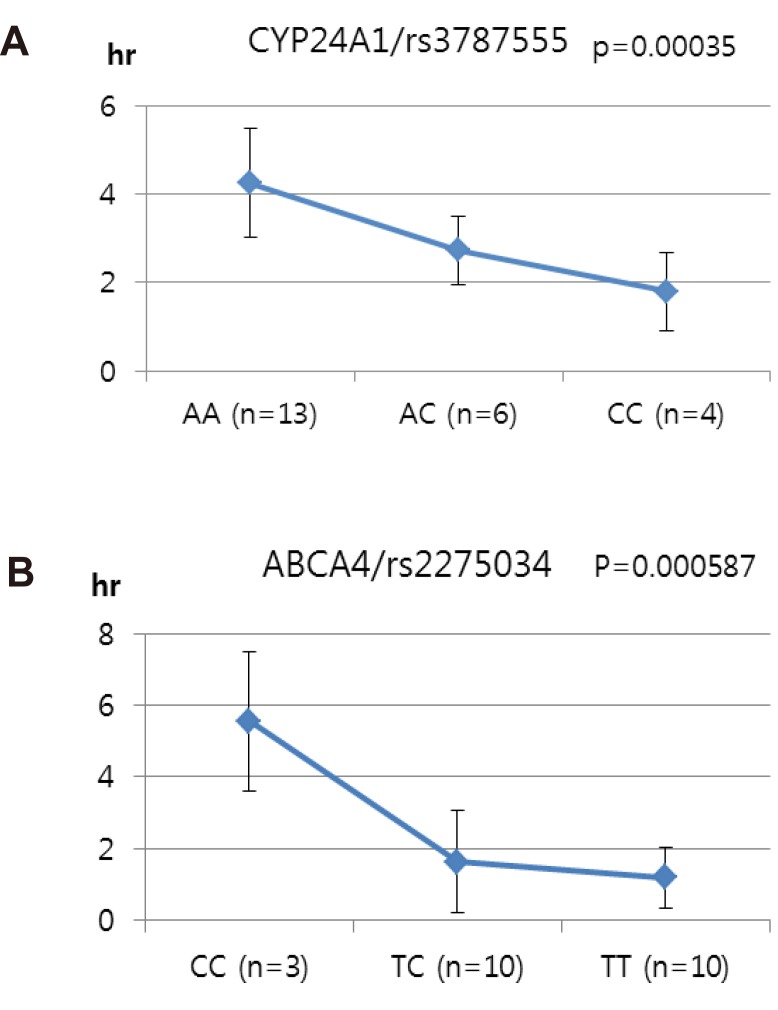
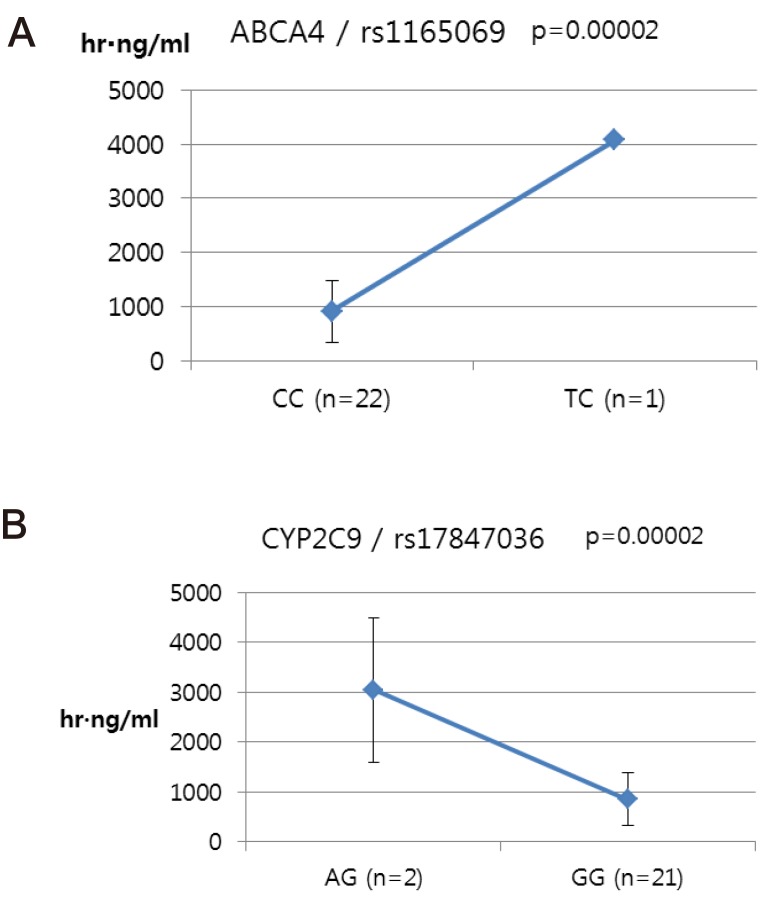
- Talniflumate is a phthalidyl ester of niflumic acid, which has potent analgesic and anti-inflammatory effects and is widely used to treat inflammatory disorders, such as rheumatoid arthritis. To screen the possible genetic factors affecting the pharmacokinetics (PK) of talniflumate, 23 male Korean volunteers were enrolled from two separate bioequivalence studies. All subjects received 740 mg (two tablets) talniflumate in a standard 2×2 cross-over model in a randomized order. For the genetic study, PK parameters of the reference drug were used. We used Illumina Human610Quad v1.0 DNA Analysis BeadChip for whole genome single nucleotide polymorphism (SNP) analysis and whole genome genotyping data were processed by linear regression analysis for PK parameters. Whole genome analysis revealed 1498 significant SNPs (P < 0.0001) for Cmax, 65 significant SNPs (P < 0.0001) for T(max), and 1491 significant SNPs (P < 0.0001) for AUC(inf). For clinical pharmacological purposes, we selected SNPs from drug metabolizing enzymes and transporters, and analyzed the PK parameters of various genotypes. Two SNPs (rs11165069 from ABCA4 (p=0.00002); rs17847036 from CYP2C9 (p=0.000001)) showed significant associations with talniflumate C(max). In the T(max) group, two SNPs (rs3787555 from CYP24A1 (p=0.00035); rs2275034 from ABCA4 (p=0.000587)) showed significant associations with talniflumate T(max). In the AUC(inf) group, two SNPs (rs11165069 from ABCA4 (p=0.00002); rs12461006 from SLC1A6 (p=0.00008)) exhibited significant associations with talniflumate absorption. These results show that genetic factors could affect the PK parameters, and provide information that may be used in the development of personalized talniflumate therapy.
Keyword
MeSH Terms
-
Absorption
Arthritis, Rheumatoid
Cytochrome P-450 CYP2C9
DNA
Genome
Genotype
Humans
Linear Models
Male
Mass Screening*
Niflumic Acid
Pharmacogenetics
Pharmacokinetics*
Polymorphism, Genetic*
Polymorphism, Single Nucleotide
Therapeutic Equivalency
Vitamin D3 24-Hydroxylase
Volunteers
Cytochrome P-450 CYP2C9
DNA
Niflumic Acid
Vitamin D3 24-Hydroxylase
Figure
Reference
-
1. Lee HW, Won KJ, Cho SH, Ha YH, Park WS, Yim HT, et al. Quantitation of niflumic acid in human plasma by high-performance liquid chromatography with ultraviolet absorbance detection and its application to a bioequivalence study of talniflumate tablets. J Chromatogr B Analyt Technol Biomed Life Sci. 2005; 821:215–220.
Article2. Knight D. Talniflumate (Genaera). Curr Opin Investig Drugs. 2004; 5:557–562.3. Lan SJ, Chando TJ, Weliky I, Schreiber EC. Metabolism of niflumic acid-14C: absorption, excretion and biotransformation by human and dog. J Pharmacol Exp Ther. 1973; 186:323–330. PMID: 4719785.4. Los M, Boned JE, Piccininali C. New esters of substituted anilinonicotinic and phenylanthranilic acids. Farmaco Sci. 1981; 36:372–385. PMID: 7238853.5. Evans WE, McLeod HL. Pharmacogenomics--drug disposition, drug targets, and side effects. N Engl J Med. 2003; 348:538–549. PMID: 12571262.6. Voisey J, Morris CP. SNP technologies for drug discovery: a current review. Curr Drug Discov Technol. 2008; 5:230–235. PMID: 18690891.
Article7. Gardiner SJ, Begg EJ. Pharmacogenetics, drug-metabolizing enzymes, and clinical practice. Pharmacol Rev. 2006; 58:521–590. PMID: 16968950.
Article8. Iyer KR, Sinz MW. Characterization of Phase I and Phase II hepatic drug metabolism activities in a panel of human liver preparations. Chem Biol Interact. 1999; 118:151–169. PMID: 10359459.
Article9. McLean C, Wilson A, Kim RB. Impact of Transporter Polymorphisms on Drug Development: Is It Clinically Significant? J Clin Pharmacol. 2016; 56(Suppl 7):S40–S58. DOI: 10.1002/jcph.691. PMID: 27385178.
Article10. Daly TM, Dumaual CM, Miao X, Farmen MW, Njau RK, Fu DJ, et al. Multiplex assay for comprehensive genotyping of genes involved in drug metabolism, excretion, and transport. Clin Chem. 2007; 53:1222–1230. PMID: 17510302.
Article11. Im S, Kim BH, Lee K, Kwack K, Yim SV. Screening study for genetic polymorphisms affecting pharmacokinetics of simvastatin. Transl Clin Pharmacol. 2016; 24:43–54.
Article12. Yun JY, Kim BH, Lee JH, Lee K, Kwack K, Yim SV. Screening study for genetic polymorphisms affecting pharmacokinetics of pioglitazone. Transl Clin Pharmacol. 2016; 24:194–202.
Article13. Mizuno N, Niwa T, Yotsumoto Y, Sugiyama Y. Impact of drug transporter studies on drug discovery and development. Pharmacol Rev. 2003; 55:425–461. PMID: 12869659.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Screening study for genetic polymorphisms affecting pharmacokinetics of simvastatin
- Role of the ABCB1 Drug Transporter Polymorphisms in the Pharmacokinetics of Oseltamivir in Humans: a Preliminary Report
- Screening study for genetic polymorphisms affecting pharmacokinetics of pioglitazone
- Tailored Pharmacotherapy of Psychotropic Drugs
- Effects of genetic polymorphisms of apolipoprotein A1 on serum HDL cholesterol level in postmenopausal Korean women