Korean J Urol.
2012 Mar;53(3):200-205.
DNA Methylation of GSTP1 in Human Prostate Tissues: Pyrosequencing Analysis
- Affiliations
-
- 1Department of Urology, Chungbuk National University College of Medicine, Cheongju, Korea. urokyj@cbnu.ac.kr
Abstract
- PURPOSE
DNA methylation is an important epigenetic mechanism of gene regulation and plays essential roles in tumor initiation and progression. Differences in methylation patterns between neoplastic and normal cells can be used to detect the presence of cancer. The aim of the present study was to evaluate the usefulness of glutathione-S-transferase-Pi (GSTP1) hypermethylation in discriminating between normal and prostate cancer (PCa) cells and in predicting tumor characteristics by use of quantitative pyrosequencing analysis.
MATERIALS AND METHODS
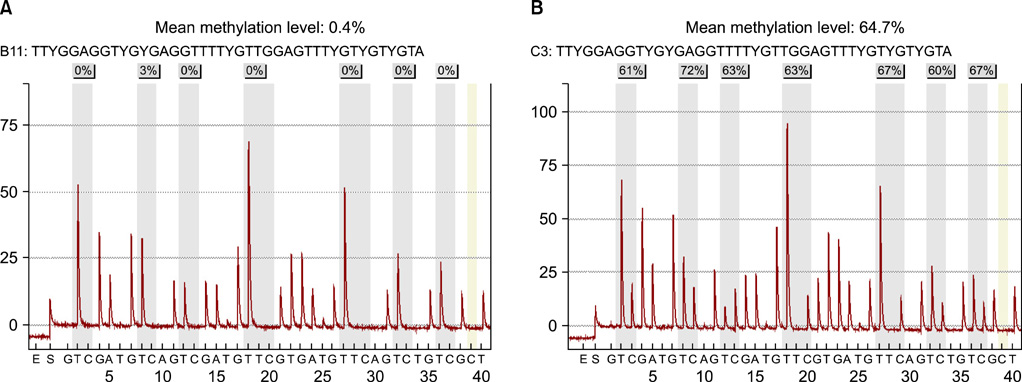
A total of 100 human prostate tissues obtained from our institute were used in this study: 45 for benign prostatic hyperplasia (BPH) and 55 for PCa. The methylation level of GSTP1 was examined by a quantitative pyrosequencing analysis. The associations between GSTP1 methylation level and clinico-pathological parameter were also compared.
RESULTS
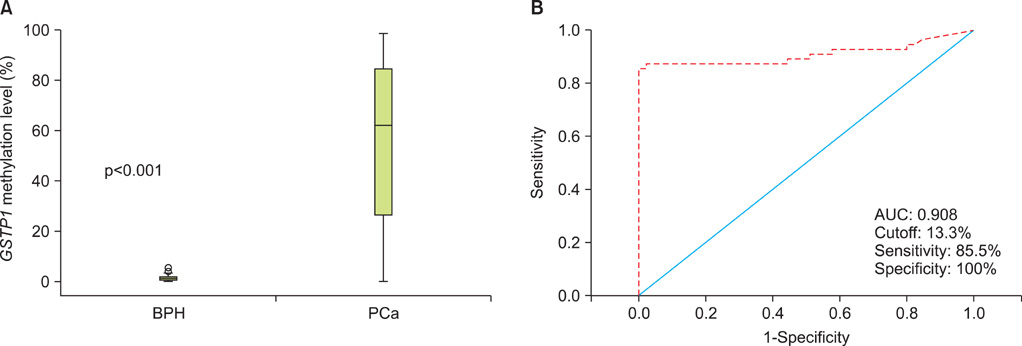
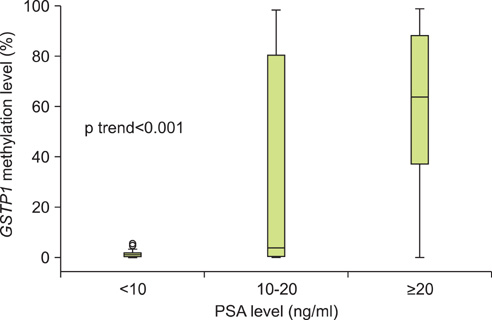
The level of GSTP1 methylation was significantly higher in PCa samples than in BPH samples (56.7+/-32.7% vs. 1.6+/-2.2%, p<0.001). The sensitivity and specificity of GSTP1 methylation status in discriminating between PCa and BPH reached 85.5% and 100%, respectively. Even after stratification by stage, Gleason score, and prostate-specific antigen (PSA) level, similar results were obtained. A positive correlation between GSTP1 methylation level and serum PSA level was observed (r=0.303, p=0.002). There were no associations between GSTP1 methylation level and age, Gleason score, and staging.
CONCLUSIONS
Our study demonstrates that GSTP1 methylation is associated with the presence of PCa and PSA levels. This methylation marker is a potentially useful indicator for the detection and monitoring of PCa.
Keyword
MeSH Terms
Figure
Reference
-
1. Greene KL, Albertsen PC, Babaian RJ, Carter HB, Gann PH, Han M, et al. Prostate specific antigen best practice statement: 2009 update. J Urol. 2009. 182:2232–2241.2. Schröder F, Kattan MW. The comparability of models for predicting the risk of a positive prostate biopsy with prostate-specific antigen alone: a systematic review. Eur Urol. 2008. 54:274–290.3. Kim HW, Ko YH, Kang SH, Lee JG. Predictive factors for prostate cancer in biopsy of patients with prostate-specific antigen levels equal to or less than 4 ng/ml. Korean J Urol. 2011. 52:166–171.4. Seo HS, Lee NK. Predictors of PSA screening among men over 40 years of age who had ever heard about PSA. Korean J Urol. 2010. 51:391–397.5. Yang JB, Jeong BC, Seo SI, Jeon SS, Choi HY, Lee HM. Outcome of prostate biopsy in men younger than 40 years of age with high prostate-specific antigen (PSA) levels. Korean J Urol. 2010. 51:21–24.6. van Vlodrop IJ, Niessen HE, Derks S, Baldewijns MM, van Criekinge W, Herman JG, et al. Analysis of promoter CpG island hypermethylation in cancer: location, location, location! Clin Cancer Res. 2011. 17:4225–4231.7. Hoque MO. DNA methylation changes in prostate cancer: current developments and future clinical implementation. Expert Rev Mol Diagn. 2009. 9:243–257.8. Ushijima T. Detection and interpretation of altered methylation patterns in cancer cells. Nat Rev Cancer. 2005. 5:223–231.9. Kim WJ, Kim YJ. Epigenetic biomarkers in urothelial bladder cancer. Expert Rev Mol Diagn. 2009. 9:259–269.10. Kim YJ, Yoon HY, Kim SK, Kim YW, Kim EJ, Kim IY, et al. EFEMP1 as a novel DNA methylation marker for prostate cancer: array-based DNA methylation and expression profiling. Clin Cancer Res. 2011. 17:4523–4530.11. Marsh S. Pyrosequencing applications. Methods Mol Biol. 2007. 373:15–24.12. Vasiljevic NA, Wu K, Brentnall AR, Kim DC, Thorat MA, Kudahetti SC, et al. Absolute quantitation of DNA methylation of 28 candidate genes in prostate cancer using pyrosequencing. Dis markers. 2011. 30:151–161.13. Phe V, Cussenot O, Roupret M. Methylated genes as potential biomarkers in prostate cancer. BJU Int. 2010. 105:1364–1370.14. Hopkins TG, Burns PA, Routledge MN. DNA methylation of GSTP1 as biomarker in diagnosis of prostate cancer. Urology. 2007. 69:11–16.15. Gonzalgo ML, Nakayama M, Lee SM, De Marzo AM, Nelson WG. Detection of GSTP1 methylation in prostatic secretions using combinatorial MSP analysis. Urology. 2004. 63:414–418.16. Alumkal JJ, Zhang Z, Humphreys EB, Bennett C, Mangold LA, Carducci MA, et al. Effect of DNA methylation on identification of aggressive prostate cancer. Urology. 2008. 72:1234–1239.17. Ellinger J, Bastian PJ, Jurgan T, Biermann K, Kahl P, Heukamp LC, et al. CpG island hypermethylation at multiple gene sites in diagnosis and prognosis of prostate cancer. Urology. 2008. 71:161–167.18. Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, et al. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004. 64:1975–1986.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Pyrosequencing Analysis of APC Methylation Level in Human Prostate Tissues: A Molecular Marker for Prostate Cancer
- Hypermethylation of Tumor-related Genes in Genitourinary Cancer Cell Lines
- DNA Methylation in Development
- A Consideration of MGMT Gene Promotor Methylation Analysis for Glioblastoma Using Methylation-Specific Polymerase Chain Reaction and Pyrosequencing
- Promoter Methylation of CDKN2A, RARbeta, and RASSF1A in Non-Small Cell Lung Carcinoma: Quantitative Evaluation Using Pyrosequencing