Ann Surg Treat Res.
2014 Dec;87(6):290-297. 10.4174/astr.2014.87.6.290.
MicroRNA expression profiling of diagnostic needle aspirates from surgical pancreatic cancer specimens
- Affiliations
-
- 1Department of Surgery, The Catholic University of Korea, Seoul St. Mary's Hospital, Seoul, Korea.
- 2Department of Surgery, The Catholic University of Korea, Bucheon St. Mary's Hospital, Bucheon, Korea. parkiy@catholic.ac.kr
- KMID: 2167084
- DOI: http://doi.org/10.4174/astr.2014.87.6.290
Abstract
- PURPOSE
MicroRNAs (miRNAs) have been widely investigated as potential biomarkers for several malignancies. To establish the feasibility of miRNA expression profiling of small biopsy samples of pancreatic cancers, we assessed expression profiles in freshly collected aspirates obtained immediately after surgical resection of the pancreas.
METHODS
We used separate fine needles (20-23 gauge) to aspirate the pancreatic cancer and adjacent normal pancreatic tissue. miRNAs that were differentially expressed in pancreatic cancers and matched paraneoplastic normal pancreatic tissues were identified using an miRNA microarray.
RESULTS
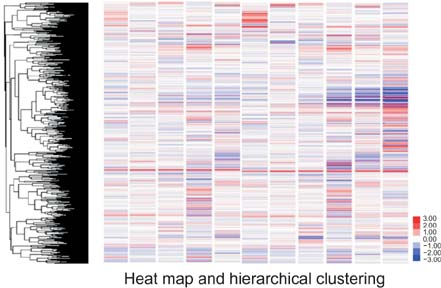
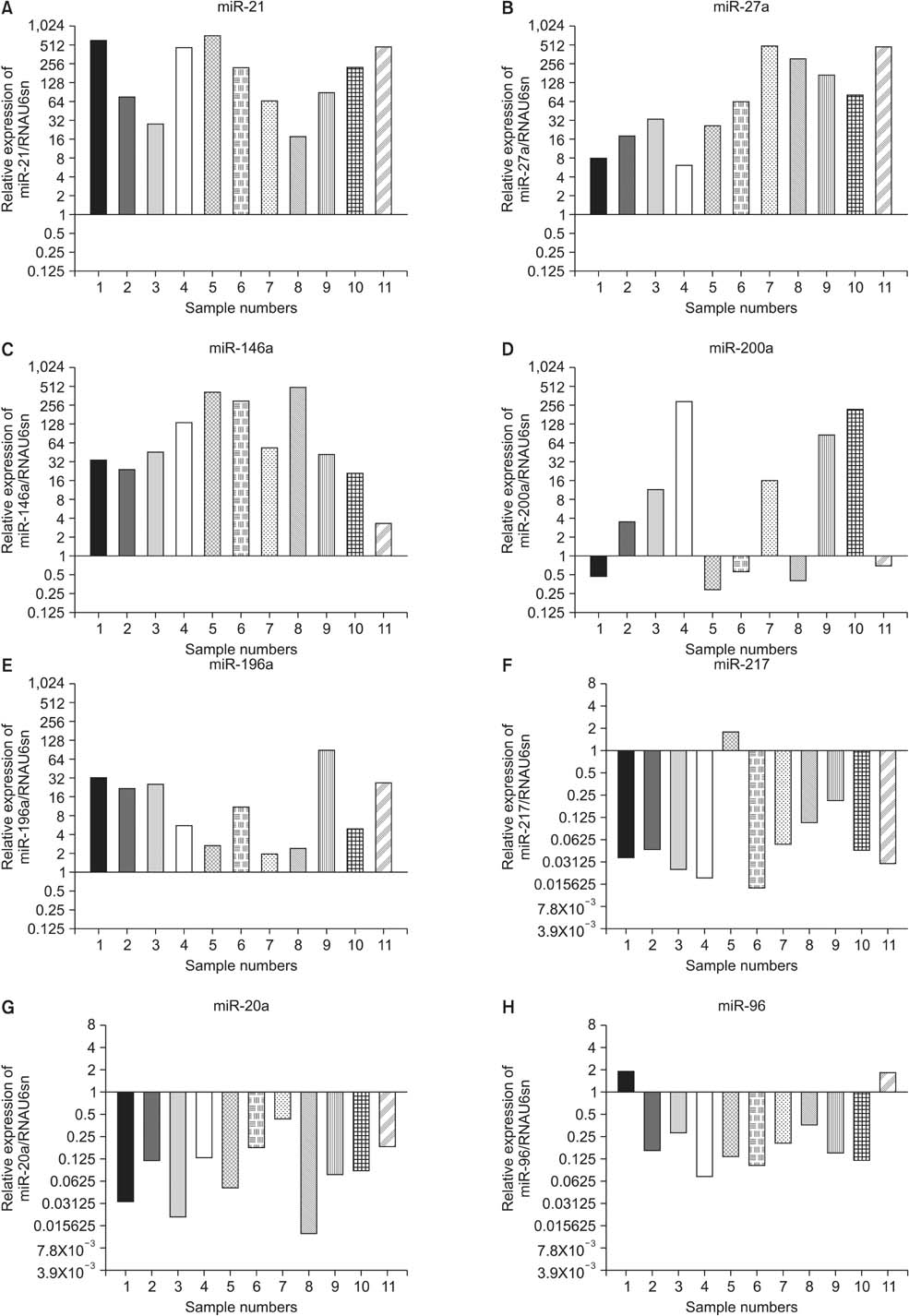
We identified 158 aberrantly expressed miRNAs in pancreatic cancers; 51 were overexpressed and 107 underexpressed compared with normal pancreatic tissue. To confirm the microarray findings, quantitative RT-PCR was performed on individual samples. We chose eight miRNAs for further analysis; of which five were overexpressed (miR-21, miR-27a, miR-146a, miR-200a, and miR-196a) and three underexpressed (miR-217, miR-20a, and miR-96) in pancreatic cancer samples compared to benign pancreatic tissue. Expression of miR-21, miR-27a, miR-146a, miR-200a, and miR-196a was significantly increased in cancer fine-needle aspirates relative to matched controls in all samples. Expression of miR-217, miR-20a, and miR-96 was significantly downregulated in almost all pancreatic cancer tissues.
CONCLUSION
We demonstrate the feasibility of performing miRNA profiling on very small specimens obtained using fine-needle aspiration of pancreatic cancers.
Keyword
MeSH Terms
Figure
Reference
-
1. Grobmyer SR, Pieracci FM, Allen PJ, Brennan MF, Jaques DP. Defi ning morbidity after pancreaticoduodenectomy: use of a prospective complication grading system. J Am Coll Surg. 2007; 204:356–364.2. Godfrey EM, Rushbrook SM, Carroll NR. Endoscopic ultrasound: a review of current diagnostic and therapeutic applications. Postgrad Med J. 2010; 86:346–353.3. Tadic M, Stoos-Veic T, Vukelic-Markovic M, Curic J, Banic M, Cabrijan Z, et al. Endoscopic ultrasound in solid pancreatic masses: current state and review of the literature. Coll Antropol. 2010; 34:337–340.4. Garcea G, Neal CP, Pattenden CJ, Steward WP, Berry DP. Molecular prognostic markers in pancreatic cancer: a systematic review. Eur J Cancer. 2005; 41:2213–2236.5. Kloosterman WP, Plasterk RH. The diverse functions of microRNAs in animal development and disease. Dev Cell. 2006; 11:441–450.6. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006; 6:857–866.7. Budhu A, Ji J, Wang XW. The clinical potential of microRNAs. J Hematol Oncol. 2010; 3:37.8. Srivastava SK, Bhardwaj A, Leavesley SJ, Grizzle WE, Singh S, Singh AP. MicroRNAs as potential clinical biomarkers: emerging approaches for their detection. Biotech Histochem. 2013; 88:373–387.9. Nakata K, Ohuchida K, Mizumoto K, Kayashima T, Ikenaga N, Sakai H, et al. MicroRNA-10b is overexpressed in pancreatic cancer, promotes its invasiveness, and correlates with a poor prognosis. Surgery. 2011; 150:916–922.10. Habbe N, Koorstra JB, Mendell JT, Offerhaus GJ, Ryu JK, Feldmann G, et al. MicroRNA miR-155 is a biomarker of early pancreatic neoplasia. Cancer Biol Ther. 2009; 8:340–346.11. Dillhoff M, Liu J, Frankel W, Croce C, Bloomston M. MicroRNA-21 is overexpressed in pancreatic cancer and a potential predictor of survival. J Gastrointest Surg. 2008; 12:2171–2176.12. Humeau M, Torrisani J, Cordelier P. miRNA in clinical practice: pancreatic cancer. Clin Biochem. 2013; 46:933–936.13. Lee EJ, Gusev Y, Jiang J, Nuovo GJ, Lerner MR, Frankel WL, et al. Expression profiling identifies microRNA signature in pancreatic cancer. Int J Cancer. 2007; 120:1046–1054.14. Steele CW, Oien KA, McKay CJ, Jamieson NB. Clinical potential of microRNAs in pancreatic ductal adenocarcinoma. Pancreas. 2011; 40:1165–1171.15. Zhang L, Jamaluddin MS, Weakley SM, Yao Q, Chen C. Roles and mechanisms of microRNAs in pancreatic cancer. World J Surg. 2011; 35:1725–1731.16. Panarelli NC, Chen YT, Zhou XK, Kitabayashi N, Yantiss RK. MicroRNA expression aids the preoperative diagnosis of pancreatic ductal adenocarcinoma. Pancreas. 2012; 41:685–690.17. Bloomston M, Frankel WL, Petrocca F, Volinia S, Alder H, Hagan JP, et al. MicroRNA expression patterns to differentiate pancreatic adenocarcinoma from normal pancreas and chronic pancreatitis. JAMA. 2007; 297:1901–1908.18. Samuel DO, Majid Mukhtar AA, Philip IO. A diagnostic pitfall: pancreatic tuberculosis, not pancreatic cancer. J Coll Physicians Surg Pak. 2013; 23:211–213.19. Sun GF, Zuo CJ, Shao CW, Wang JH, Zhang J. Focal autoimmune pancreatitis: radiological characteristics help to distinguish from pancreatic cancer. World J Gastroenterol. 2013; 19:3634–3641.20. Matsumoto I, Shinzeki M, Toyama H, Asari S, Goto T, Yamada I, et al. A focal mass-forming autoimmune pancreatitis mimicking pancreatic cancer with obstruction of the main pancreatic duct. J Gastrointest Surg. 2011; 15:2296–2298.21. van Gulik TM, Reeders JW, Bosma A, Moojen TM, Smits NJ, Allema JH, et al. Incidence and clinical findings of benign, inflammatory disease in patients resected for presumed pancreatic head cancer. Gastrointest Endosc. 1997; 46:417–423.22. Jiao LR, Frampton AE, Jacob J, Pellegrino L, Krell J, Giamas G, et al. MicroRNAs targeting oncogenes are down-regulated in pancreatic malignant transformation from benign tumors. PLoS One. 2012; 7:e32068.23. Singh P, Srinivasan R, Wig JD. Major molecular markers in pancreatic ductal adenocarcinoma and their roles in screening, diagnosis, prognosis, and treatment. Pancreas. 2011; 40:644–652.24. Toiyama Y, Hur K, Tanaka K, Inoue Y, Kusunoki M, Boland CR, et al. Serum miR-200c is a novel prognostic and metastasispredictive biomarker in patients with colorectal cancer. Ann Surg. 2014; 259:735–743.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Diagnostic Utility of K-ras Mutation in the Sample of Percutaneous Needle Aspiraton of Pancreatic Cancer and Chronic Pancreatitis
- The Utilization of Cytologic Fine-Needle Aspirates of Lung Cancer for Molecular Diagnostic Testing
- Clinical utility of endoscopic ultrasound-guided tissue acquisition for comprehensive genomic profiling of pancreatic cancer
- Cytologic Classification of Fibrocystic Disease of the Breast: A Proposal for Use of Cytologic Criteria Grading System
- Clinical utility of endoscopic ultrasound-guided tissue acquisition for comprehensive genomic profiling of patients with biliary tract cancer, especially with intrahepatic cholangiocarcinoma