Allergy Asthma Immunol Res.
2015 Sep;7(5):449-457. 10.4168/aair.2015.7.5.449.
Altered microRNA Expression Profiles of Extracellular Vesicles in Nasal Mucus From Patients With Allergic Rhinitis
- Affiliations
-
- 1Department of Otolaryngology, First People Hospital of Zhangjiagang City, Suzhou, China. Gordon-wu@qq.com
- 2Department of Otolaryngology, Huadong Hospital, Fudan University, Shanghai, China.
- 3Institute of Neuroscience and Department of Neurobiology and Psychology, Key lab of Pain Research and Therapy, Soochow University, Suzhou, China.
- 4Department of Science and Education, First People Hospital of Zhangjiagang City, Suzhou, China.
- 5Department of Otolaryngology, Changhai Hospital, Second Military Medical University, Shanghai, China.
- 6Department of Thoracic Surgery, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan, China.
- KMID: 2147953
- DOI: http://doi.org/10.4168/aair.2015.7.5.449
Abstract
- PURPOSE
Allergic rhinitis (AR) is an inflammatory disorder of the upper airway. Exosomes or extracellular vesicles are nanosized vesicles of endosomal origin released from inflammatory and epithelial cells that have been implicated in allergic diseases. In this study, we characterized the microRNA (miRNA) content of exosomes in AR.
METHODS
Extracellular vesicles were isolated from nasal mucus from healthy control subjects (n=10) and patients with severe AR (n=10). Vesicle RNA was analyzed by using a TaqMan microRNA assays Human Panel-Early Access kit (Applied Biosystems, Foster City, CA, USA) containing probes for 366 human miRNAs, and selected findings were validated with quantitative RT-PCR. Target prediction and pathway analysis for the differentially expressed miRNAs were performed using DIANA-mirPath.
RESULTS
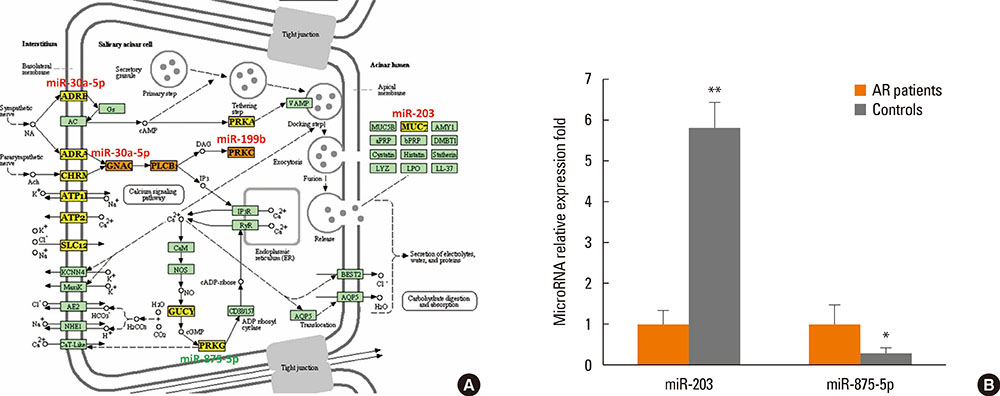
Twenty-one vesicle miRNAs were up-regulated and 14 miRNAs were under-regulated significantly (P<0.05) in nasal mucus from AR patients when compared to healthy controls. Bioinformatic analysis by DIANA-mirPath demonstrated that 32 KEGG biological processes were significantly enriched (P<0.05, FDR corrected) among differentially expressed vesicle miRNA signatures. Among them, the B-cell receptor signaling pathway (P=3.709E-09), the natural killer cell-mediated cytotoxicity (P=8.466E-05), the T-cell receptor signaling pathway (P=0.00075), the RIG-I-like receptor signaling pathway (P=0.00127), the Wnt signaling pathway (P=0.00130), endocytosis (P=0.00440), and salivary secretion (P=0.04660) were the most prominent pathways enriched in quantiles with differential vesicle miRNA patterns. Furthermore, miR-30-5p, miR-199b-3p, miR-874, miR-28-3p, miR-203, and miR-875-5p, involved in B-cell receptor and salivary secretion signaling pathways, were selected for validation using independent samples from 44 AR patients and 20 healthy controls. MiR-30-5p and miR-199b-3p were significantly increased in extracellular vesicles from nasal mucus when compared to healthy controls, while miR-874 and miR-28-3p were significantly down-regulated. In addition, miRNA-203 was significantly increased in AR patients, while miRNA-875-5p was found to be significantly decreased in AR patients.
CONCLUSIONS
This study demonstrated that vesicle miRNA may be a regulator for the development of AR.
Keyword
MeSH Terms
Figure
Reference
-
1. Bousquet J, Khaltaev N, Cruz AA, Denburg J, Fokkens WJ, Togias A, et al. Allergic Rhinitis and its Impact on Asthma (ARIA) 2008 update (in collaboration with the World Health Organization, GA(2)LEN and AllerGen). Allergy. 2008; 63:Suppl 86. 8–160.2. Aït-Khaled N, Pearce N, Anderson HR, Ellwood P, Montefort S, Shah J, et al. Global map of the prevalence of symptoms of rhinoconjunctivitis in children: the International Study of Asthma and Allergies in Childhood (ISAAC) Phase Three. Allergy. 2009; 64:123–148.3. International Rhinitis Management Working Group. International Consensus Report on the diagnosis and management of rhinitis. Allergy. 1994; 49:1–34.4. van Niel G, Raposo G, Candalh C, Boussac M, Hershberg R, Cerf-Bensussan N, et al. Intestinal epithelial cells secrete exosome-like vesicles. Gastroenterology. 2001; 121:337–349.5. Admyre C, Johansson SM, Qazi KR, Filén JJ, Lahesmaa R, Norman M, et al. Exosomes with immune modulatory features are present in human breast milk. J Immunol. 2007; 179:1969–1978.6. Michael A, Bajracharya SD, Yuen PS, Zhou H, Star RA, Illei GG, et al. Exosomes from human saliva as a source of microRNA biomarkers. Oral Dis. 2010; 16:34–38.7. Lässer C, Alikhani VS, Ekström K, Eldh M, Paredes PT, Bossios A, et al. Human saliva, plasma and breast milk exosomes contain RNA: uptake by macrophages. J Transl Med. 2011; 9:9.8. Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ, Lötvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007; 9:654–659.9. Yuan A, Farber EL, Rapoport AL, Tejada D, Deniskin R, Akhmedov NB, et al. Transfer of microRNAs by embryonic stem cell microvesicles. PLoS One. 2009; 4:e4722.10. Zomer A, Vendrig T, Hopmans ES, van Eijndhoven M, Middeldorp JM, Pegtel DM. Exosomes: fit to deliver small RNA. Commun Integr Biol. 2010; 3:447–450.11. Ratajczak J, Miekus K, Kucia M, Zhang J, Reca R, Dvorak P, et al. Embryonic stem cell-derived microvesicles reprogram hematopoietic progenitors: evidence for horizontal transfer of mRNA and protein delivery. Leukemia. 2006; 20:847–856.12. Ratajczak J, Wysoczynski M, Hayek F, Janowska-Wieczorek A, Ratajczak MZ. Membrane-derived microvesicles: important and underappreciated mediators of cell-to-cell communication. Leukemia. 2006; 20:1487–1495.13. Mack M, Kleinschmidt A, Brühl H, Klier C, Nelson PJ, Cihak J, et al. Transfer of the chemokine receptor CCR5 between cells by membrane-derived microparticles: a mechanism for cellular human immunodeficiency virus 1 infection. Nat Med. 2000; 6:769–775.14. Suojalehto H, Lindström I, Majuri ML, Mitts C, Karjalainen J, Wolff H, et al. Altered microRNA expression of nasal mucosa in long-term asthma and allergic rhinitis. Int Arch Allergy Immunol. 2014; 163:168–178.15. Zhang XH, Zhang YN, Liu Z. MicroRNA in chronic rhinosinusitis and allergic rhinitis. Curr Allergy Asthma Rep. 2014; 14:415.16. The European Academy of Allergology and Clinical Immunology. Position paper: allergen standardization and skin tests. Allergy. 1993; 48:48–82.17. Ruocco L, Fattori B, Romanelli A, Martelloni M, Casani A, Samolewska M, et al. A new collection method for the evaluation of nasal mucus proteins. Clin Exp Allergy. 1998; 28:881–888.18. Jansen FH, Krijgsveld J, van Rijswijk A, van den Bemd GJ, van den Berg MS, van Weerden WM, et al. Exosomal secretion of cytoplasmic prostate cancer xenograft-derived proteins. Mol Cell Proteomics. 2009; 8:1192–1205.19. Johansson SM, Admyre C, Scheynius A, Gabrielsson S. Different types of in vitro generated human monocyte-derived dendritic cells release exosomes with distinct phenotypes. Immunology. 2008; 123:491–499.20. Anglicheau D, Sharma VK, Ding R, Hummel A, Snopkowski C, Dadhania D, et al. MicroRNA expression profiles predictive of human renal allograft status. Proc Natl Acad Sci U S A. 2009; 106:5330–5335.21. Papadopoulos GL, Alexiou P, Maragkakis M, Reczko M, Hatzigeorgiou AG. DIANA-mirPath: integrating human and mouse microRNAs in pathways. Bioinformatics. 2009; 25:1991–1993.22. Reczko M, Maragkakis M, Alexiou P, Grosse I, Hatzigeorgiou AG. Functional microRNA targets in protein coding sequences. Bioinformatics. 2012; 28:771–776.23. Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, et al. DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res. 2013; 41:W169–W173.24. Hsu SD, Lin FM, Wu WY, Liang C, Huang WC, Chan WL, et al. miR-TarBase: a database curates experimentally validated microRNA-target interactions. Nucleic Acids Res. 2011; 39:D163–D169.25. Scian MJ, Maluf DG, David KG, Archer KJ, Suh JL, Wolen AR, et al. MicroRNA profiles in allograft tissues and paired urines associate with chronic allograft dysfunction with IF/TA. Am J Transplant. 2011; 11:2110–2122.26. Zhang Y, Zhang L. Prevalence of allergic rhinitis in china. Allergy Asthma Immunol Res. 2014; 6:105–113.27. Dhong HJ. Classification of allergic rhinitis: what is most suitable in Korea? Allergy Asthma Immunol Res. 2013; 5:65–67.28. Booth AM, Fang Y, Fallon JK, Yang JM, Hildreth JE, Gould SJ. Exosomes and HIV Gag bud from endosome-like domains of the T cell plasma membrane. J Cell Biol. 2006; 172:923–935.29. Balaj L, Lessard R, Dai L, Cho YJ, Pomeroy SL, Breakefield XO, et al. Tumour microvesicles contain retrotransposon elements and amplified oncogene sequences. Nat Commun. 2011; 2:180.30. Li XB, Zhang ZR, Schluesener HJ, Xu SQ. Role of exosomes in immune regulation. J Cell Mol Med. 2006; 10:364–375.31. Shaoqing Y, Ruxin Z, Guojun L, Zhiqiang Y, Hua H, Shudong Y, et al. Microarray analysis of differentially expressed microRNAs in allergic rhinitis. Am J Rhinol Allergy. 2011; 25:e242–e246.32. Suojalehto H, Toskala E, Kilpeläinen M, Majuri ML, Mitts C, Lindström I, et al. MicroRNA profiles in nasal mucosa of patients with allergic and nonallergic rhinitis and asthma. Int Forum Allergy Rhinol. 2013; 3:612–620.33. Chen RF, Huang HC, Ou CY, Hsu TY, Chuang H, Chang JC, et al. MicroRNA-21 expression in neonatal blood associated with antenatal immunoglobulin E production and development of allergic rhinitis. Clin Exp Allergy. 2010; 40:1482–1490.34. Lee YN, Tuckerman J, Nechushtan H, Schutz G, Razin E, Angel P. c-Fos as a regulator of degranulation and cytokine production in FcepsilonRI-activated mast cells. J Immunol. 2004; 173:2571–2577.35. Odom S, Gomez G, Kovarova M, Furumoto Y, Ryan JJ, Wright HV, et al. Negative regulation of immunoglobulin E-dependent allergic responses by Lyn kinase. J Exp Med. 2004; 199:1491–1502.36. Sharma P, Dudus L, Nielsen PA, Clausen H, Yankaskas JR, Hollingsworth MA, et al. MUC5B and MUC7 are differentially expressed in mucous and serous cells of submucosal glands in human bronchial airways. Am J Respir Cell Mol Biol. 1998; 19:30–37.37. Li T, Leong MH, Harms B, Kennedy G, Chen L. MicroRNA-21 as a potential colon and rectal cancer biomarker. World J Gastroenterol. 2013; 19:5615–5621.38. Siasos G, Kollia C, Tsigkou V, Basdra EK, Lymperi M, Oikonomou E, et al. MicroRNAs: novel diagnostic and prognostic biomarkers in atherosclerosis. Curr Top Med Chem. 2013; 13:1503–1517.39. Baxter D, McInnes IB, Kurowska-Stolarska M. Novel regulatory mechanisms in inflammatory arthritis: a role for microRNA. Immunol Cell Biol. 2012; 90:288–292.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Correlation Study between the Specific IgE for Staphylococcus Aureus Exotoxin and Nasal Mucus Culture in Allergic Rhinitis
- Diagnosis of Allergic Rhinitis
- Upregulation of the Vitamin D Receptor in the Nasal Mucosa of Patients With Allergic Rhinitis
- Expression of MMP-9 and TIMP-1 in the Nasal Mucosa of Allergic Rhinitis
- Evidences for Local Allergic Rhinitis