J Vet Sci.
2015 Dec;16(4):491-500. 10.4142/jvs.2015.16.4.491.
Genetic characterization of bovine viral diarrhea virus strains in Beijing, China and innate immune responses of peripheral blood mononuclear cells in persistently infected dairy cattle
- Affiliations
-
- 1College of Veterinary Medicine, China Agricultural University, Beijing 100193, China. jiufeng_wang@hotmail.com
- KMID: 2133632
- DOI: http://doi.org/10.4142/jvs.2015.16.4.491
Abstract
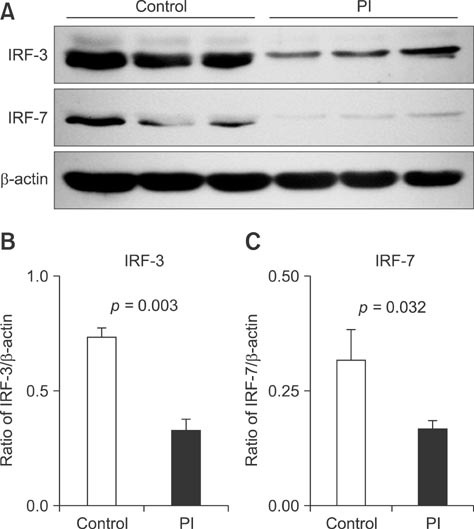
- To acquire epidemiological data on the bovine viral diarrhea virus (BVDV) and identify cattle persistently infected (PI) with this virus, 4,327 samples from Holstein dairy cows were screened over a four-year period in Beijing, China. Eighteen BVD viruses were isolated, 12 from PI cattle. Based on genetic analysis of their 5'-untranslated region (5'-UTR), the 18 isolates were assigned to subgenotype BVDV-1m, 1a, 1d, 1q, and 1b. To investigate the innate immune responses in the peripheral-blood mononuclear cells of PI cattle, the expression of Toll-like receptors (TLRs), RIG-I-like receptors, interferon-alpha (IFN-alpha), IFN-beta, myxovirus (influenza virus) resistance 1 (MX1), and interferon stimulatory gene 15 (ISG15) was assessed by qPCR. When compared with healthy cattle, the expression of TLR-7, IFN-alpha, and IFN-beta mRNA was downregulated, but the expression of MX1 and ISG-15 mRNA was upregulated in PI cattle. Immunoblotting analysis revealed that the expression of interferon regulatory factor 3 (IRF-3) and IRF-7 was lower in PI cattle than in healthy cattle. Thus, BVDV-1m and 1a are the predominant subgenotypes in the Beijing region, and the strains are highly divergent. Our findings also suggest that the TLR-7/IRF-7 signaling pathway plays a role in evasion of host restriction by BVDV.
Keyword
MeSH Terms
Figure
Reference
-
1. Baker JC. The clinical manifestations of bovine viral diarrhea infection. Vet Clin North Am Food Anim Pract. 1995; 11:425–445.
Article2. Berke IC, Yu X, Modis Y, Egelman EH. MDA5 assembles into a polar helical filament on dsRNA. Proc Natl Acad Sci U S A. 2012; 109:18437–18441.
Article3. Blasius AL, Beutler B. Intracellular toll-like receptors. Immunity. 2010; 32:305–315.
Article4. Booth RE, Thomas CJ, El-Attar LM, Gunn G, Brownlie J. A phylogenetic analysis of Bovine viral diarrhoea virus (BVDV) isolates from six different regions of the UK and links to animal movement data. Vet Res. 2013; 44:43.
Article5. Chang S, Kodys K, Szabo G. Impaired expression and function of toll-like receptor 7 in hepatitis C virus infection in human hepatoma cells. Hepatology. 2010; 51:35–42.
Article6. Deng Y, Sun CQ, Cao SJ, Lin T, Yuan SS, Zhang HB, Zhai SL, Huang L, Shan TL, Zheng H, Wen XT, Tong GZ. High prevalence of bovine viral diarrhea virus 1 in Chinese swine herds. Vet Microbiol. 2012; 159:490–493.
Article7. Fiebach AR, Guzylack-Piriou L, Python S, Summerfield A, Ruggli N. Classical swine fever virus Npro limits type I interferon induction in plasmacytoid dendritic cells by interacting with interferon regulatory factor 7. J Virol. 2011; 85:8002–8011.
Article8. Gao S, Luo J, Du J, Lang Y, Cong G, Shao J, Lin T, Zhao F, Belák S, Liu L, Chang H, Yin H. Serological and molecular evidence for natural infection of Bactrian camels with multiple subgenotypes of bovine viral diarrhea virus in Western China. Vet Microbiol. 2013; 163:172–176.
Article9. Glew EJ, Carr BV, Brackenbury LS, Hope JC, Charleston B, Howard CJ. Differential effects of bovine viral diarrhoea virus on monocytes and dendritic cells. J Gen Virol. 2003; 84:1771–1780.10. Gong X, Cao X, Zheng F, Chen Q, Zhou J, Yin H, Liu L, Cai X. Identification and characterization of a novel subgenotype of bovine viral diarrhea virus isolated from dairy cattle in Northwestern China. Virus Genes. 2013; 46:375–376.
Article11. Hanon JB, Van der Stede Y, Antonissen A, Mullender C, Tignon M, van den Berg T, Caij B. Distinction between persistent and transient infection in a bovine viral diarrhoea (BVD) control programme: appropriate interpretation of real-time RT-PCR and antigen-ELISA test results. Transbound Emerg Dis. 2014; 61:156–162.
Article12. Heuer C, Healy A, Zerbini C. Economic effects of exposure to bovine viral diarrhea virus on dairy herds in New Zealand. J Dairy Sci. 2007; 90:5428–5438.
Article13. Hilton L, Moganeradj K, Zhang G, Chen YH, Randall RE, McCauley JW, Goodbourn S. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J Virol. 2006; 80:11723–11732.
Article14. Hiscott J. Triggering the innate antiviral response through IRF-3 activation. J Biol Chem. 2007; 282:15325–15329.
Article15. Honda K, Yanai H, Negishi H, Asagiri M, Sato M, Mizutani T, Shimada N, Ohba Y, Takaoka A, Yoshida N, Taniguchi T. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005; 434:772–777.
Article16. Houe H. Epidemiological features and economical importance of bovine virus diarrhoea virus (BVDV) infections. Vet Microbiol. 1999; 64:89–107.
Article17. Houe H. Epidemiology of bovine viral diarrhea virus. Vet Clin North Am Food Anim Pract. 1995; 11:521–547.
Article18. Hüsser L, Alves MP, Ruggli N, Summerfield A. Identification of the role of RIG-I, MDA-5 and TLR3 in sensing RNA viruses in porcine epithelial cells using lentivirus-driven RNA interference. Virus Res. 2011; 159:9–16.
Article19. Iqbal M, Poole E, Goodbourn S, McCauley JW. Role for bovine viral diarrhea virus Erns glycoprotein in the control of activation of beta interferon by double-stranded RNA. J Virol. 2004; 78:136–145.
Article20. Jeon YJ, Yoo HM, Chung CH. ISG15 and immune diseases. Biochim Biophys Acta. 2010; 1802:485–496.
Article21. Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006; 7:131–137.
Article22. Lee SW, Park Y, So T, Kwon BS, Cheroutre H, Mittler RS, Croft M. Identification of regulatory functions for 4-1BB and 4-1BBL in myelopoiesis and the development of dendritic cells. Nat Immunol. 2008; 9:917–926.
Article23. Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005; 5:375–386.
Article24. Potgieter LN. Immunology of bovine viral diarrhea virus. Vet Clin North Am Food Anim Pract. 1995; 11:501–520.
Article25. Ridpath JF, Fulton RW, Kirkland PD, Neill JD. Prevalence and antigenic differences observed between Bovine viral diarrhea virus subgenotypes isolated from cattle in Australia and feedlots in the southwestern United States. J Vet Diagn Invest. 2010; 22:184–191.
Article26. Sadler AJ, Williams BRG. Interferon-inducible antiviral effectors. Nat Rev Immunol. 2008; 8:559–568.
Article27. Schmid S, Mordstein M, Kochs G, García-Sastre A, tenOever BR. Transcription factor redundancy ensures induction of the antiviral state. J Biol Chem. 2010; 285:42013–42022.
Article28. Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature. 2011; 472:481–485.
Article29. Schweizer M, Mätzener P, Pfaffen G, Stalder H, Peterhans E. "Self" and "nonself" manipulation of interferon defense during persistent infection: bovine viral diarrhea virus resists alpha/beta interferon without blocking antiviral activity against unrelated viruses replicating in its host cells. J Virol. 2006; 80:6926–6935.
Article30. Talantov D, Mazumder A, Yu JX, Briggs T, Jiang Y, Backus J, Atkins D, Wang Y. Novel genes associated with malignant melanoma but not benign melanocytic lesions. Clin Cancer Res. 2005; 11:7234–7242.
Article31. Vilcek S, Durkovic B, Kolesárová M, Greiser-Wilke I, Paton D. Genetic diversity of international bovine viral diarrhoea virus (BVDV) isolates: identification of a new BVDV-1 genetic group. Vet Res. 2004; 35:609–615.
Article32. Vilcek S, Herring AJ, Herring JA, Nettleton PF, Lowings JP, Paton DJ. Pestiviruses isolated from pigs, cattle and sheep can be allocated into at least three genogroups using polymerase chain reaction and restriction endonuclease analysis. Arch Virol. 1994; 136:309–323.
Article33. Vilcek S, Paton DJ, Durkovic B, Strojny L, Ibata G, Moussa A, Loitsch A, Rossmanith W, Vega S, Scicluna MT, Paifi V. Bovine viral diarrhoea virus genotype 1 can be separated into at least eleven genetic groups. Arch Virol. 2001; 146:99–115.
Article34. Wang X, Tu C, Li H, Jin K, Xuan H, Chang G, Sun H, Zhu W, Fei E, Yin Z. Pigs naturally infected by bovine diarrhea virus present signs resembling hog cholera. Chin J Vet Sci. 1996; 16:341–345.35. Xue F, Zhu YM, Li J, Zhu LC, Ren XG, Feng JK, Shi HF, Gao YR. Genotyping of bovine viral diarrhea viruses from cattle in China between 2005 and 2008. Vet Microbiol. 2010; 143:379–383.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Tissue distribution of bovine viral diarrhea virus antigens in persistently infected cattle
- Development of a novel diagnostic test for detection of bovine viral diarrhea persistently infected animals using hair
- Genetic Typing of Bovine Viral Diarrhea Viruses (BVDV) Circulating in Korea
- Prevalence of bovine viral diarrhea virus from Korean native cattle farms in Jeju
- Infectivity of bovine leukemia virus to Korean native goats I. antibody responses and syncytium assay for Korean native goats experimentally infected with bovine leukemia virus