J Bacteriol Virol.
2010 Mar;40(1):11-18. 10.4167/jbv.2010.40.1.11.
Quantitative and Qualitative Estimation of Bacteria Contaminating Human Hairs
- Affiliations
-
- 1Department of Beauty Arts, Seokyeong University, Seoul, Korea.
- 2Department of Biological Engineering, Seokyeong University, Seoul, Korea. baakdoo@skuniv.ac.kr
- KMID: 1456249
- DOI: http://doi.org/10.4167/jbv.2010.40.1.11
Abstract
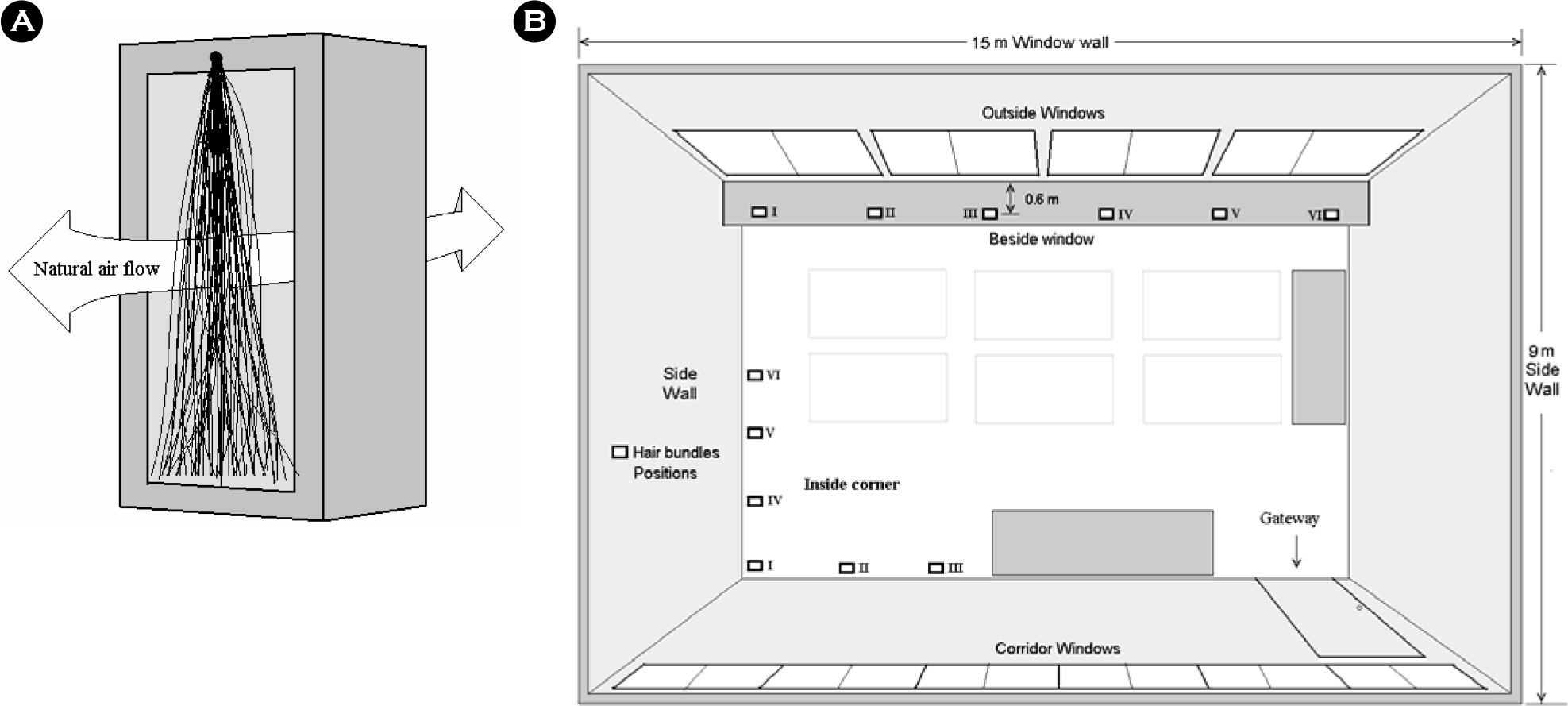
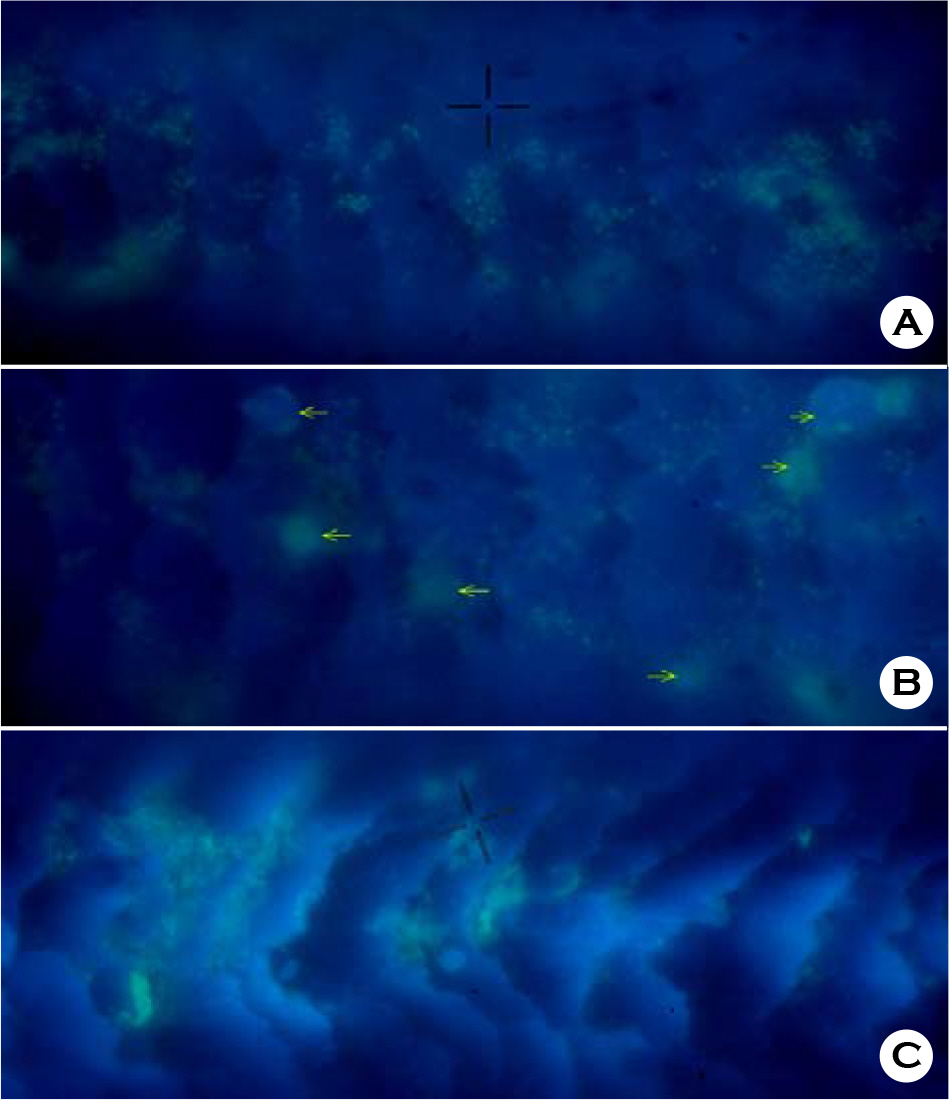
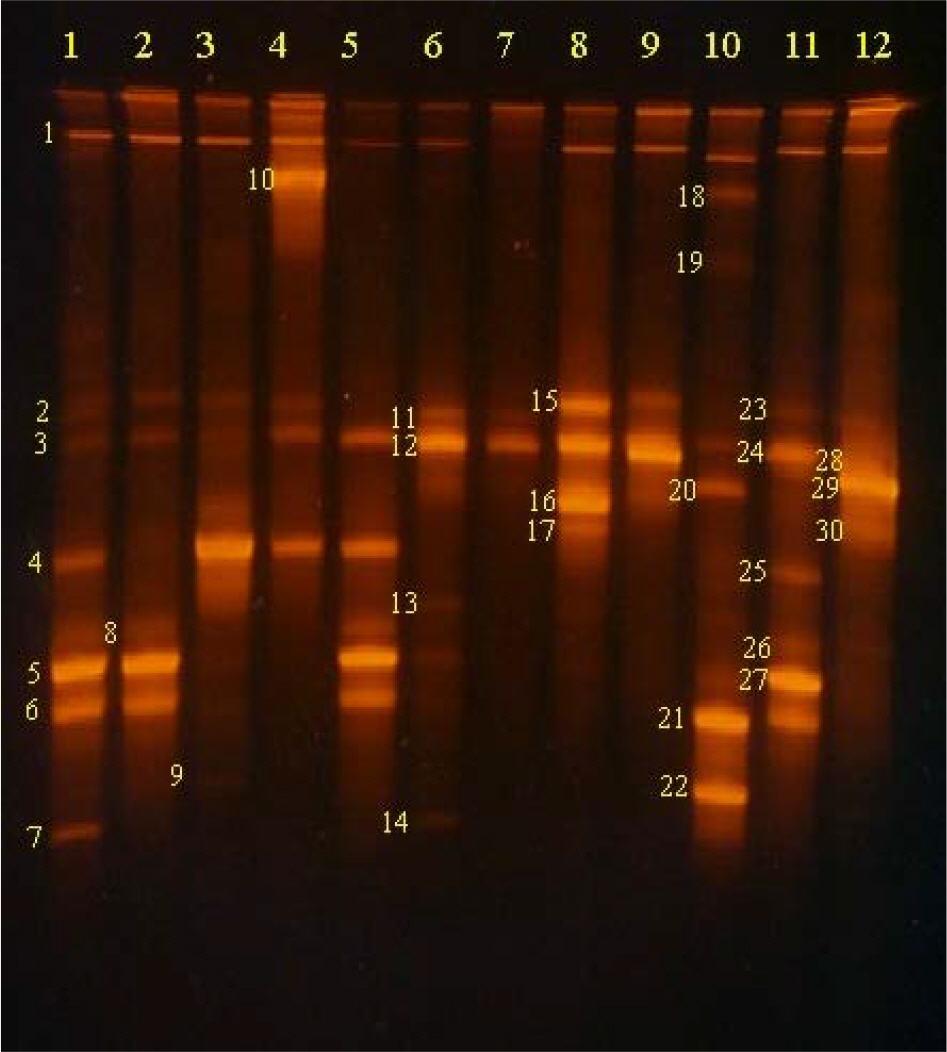
- Human hairs have been known to be easily contaminated with microorganisms. This study was performed in order to measure what bacterial species and how much microorganisms contaminate human hairs in specific place. Virgin human hairs were left at 6 positions in inside corner and beside window in a laboratory for 7 days. The number of viable bacterial cells, which were determined by most probable number method, contaminating the human hairs was measured at a maximum of 10(6)/g hair and a minimum of 10(3)/g hair in inside corner and maximum of 10(6)/g hair and a minimum of 10(3)/g hair beside window. The bacterial cells-contaminating human hairs were observed via fluorescence light microscopy after 4',6-diamino-2-phenylindole (DAPI) staining. The bacterial community contaminating human hairs was analyzed via the thermal gradient gel electrophoresis (TGGE) technique, based on the diversity of the 16S-rDNA variable region. In total, approximately 20 bacterial species were detected from 12 groups of hair samples. In this study, general experimental methods-fluorescence staining, TGGE and MPN-were combined to develop new method for observation and estimation of bacteria contaminating human hairs.
MeSH Terms
Figure
Reference
-
1). Bollin GE., Plouffe JF., Para MF., Hackman B. Aerosols containing Legionella pneumophila generated by shower heads and hot-water faucets. Appl Environ Microbiol. 1985. 50:1128–31.2). Sttar SA., Ijaz MK., Gerba CP. Spread of viral infection by aerosols. Crit Rev Environ Sci Tech. 1987. 17:89–131.3). Bernstein IL., Safferman R. Clinical sensitivity to green algae demonstrated by nasal challenge and in vitro tests of immediate hypersensitivity. J Allergy Clin Immunol. 1973. 51:22–8.4). Croft WA., Jarvis BB., Yatawara CS. Airborne outbreak of trichothecene toxicosis. Atmos Environ. 1986. 20:549–52.
Article5). Castellan RM., Olenchock SA., Kinsley KB., Hankinson JL. Inhaled endotoxin and decreased spirometric values. N Engl J Med. 1987. 317:605–10.
Article6). Jacobs RR. Airborne endotoxins: an association with occupational lung disease. Appl Ind Hyg. 1989. 4:50–6.
Article7). Flaherty DK., Deck FH., Cooper J., Bishop K., Winzenburger PA., Smith LR, et al. Bacterial endotoxin isolated from a water spray humidification system as a putative agent of occupationrelated lung disease. Infect Immun. 1984. 43:206–12.8). Samson RA. Occurrence of moulds in modern living and working environments. Eur J Epidemiol. 1985. 1:54–61.
Article9). Reynolds SJ., Striefel AJ., McJilton CE. Elevated airborne concentrations of fungi in residential and office environments. Am Ind Hyg Assoc J. 1990. 51:601–4.
Article10). Thorne PS., Kiekhaefer MS., Whitten P., Donham KJ. Comparison of bioaerosol sampling methods in barns housing swine. Appl Environ Microbiol. 1992. 58:2543–51.
Article11). Greene VW., Vesley D., Bond RG., Michaelsen GS. Microbiological contamination of hospital air. I. Quantitative studies. Appl Environ Microbiol. 1962. 10:561–6.12). Favoero MS., Puleo JR., Marshall JH., Oxborrow GS. Comparative levels and types of microbial contamination detected in industrial clean rooms. Appl Microbiol. 1966. 14:539–51.
Article13). Favoero MS., Puleo JR., Marshall JH., Oxborrow GS. Comparison of microbial contamination levels among hospital operating room and industrial clean rooms. Appl Microbiol. 1968. 16:480–6.14). Greene VW., Vesley D., Bond RG., Michaelsen GS. Microbiological contamination of hospital air. II. Qualitative studies. Appl Environ Microbiol. 1962. 10:567–71.15). Hall LB. Air sampling for hospitals. Hospital Topics Magazine. 1962. 40:97–100.16). Sperling LC. Hair anatomy for the clinician. J Am Acad Dermatol. 1991. 25:1–17.
Article17). Ward DM., Weller R., Bateson MM. 16S rRNA sequences reveal numerous uncultured microorganisms in a natural community. Nature. 1990. 345:63–5.
Article18). Spratt DA., Weightman AJ., Wade WG. Diversity of oral asaccharolytic Eubacterium species in periodontitis-identification of novel phylotypes representing uncultivated taxa. Oral Microbiol Immunol. 1999. 14:56–9.19). Headington JT. Transverse microscopic anatomy of the human scalp. A basis for a morphometric approach to disorders of the hair follicle. Arch Dermatol. 1984. 120:449–56.
Article20). Kim SH. A study on characteristics and amino acid composition of female hair according to age period. J Kor Soc Cosm. 2007. 13:1111–1120.21). de Lacharriere O., Deloche C., Misciali C., Piraccini BM., Vincenzi C., Bastien P, et al. Hair diameter diversity: a clinical sign reflecting the follicle miniaturization. Arch Dermatol. 2001. 137:641–6.22). Greenberg AE., Clesceri LS., Eaton AD., Franson MAH. Standard methods for the examination of water and wastewater. 18th ed. Washington DC; American Public Health Association. 1992. pp.9-49~. 9–50.23). Saby S., Sibille I., Mathieu L., Paquin JL., Block JC. Influence of water chlorination on the counting of bacteria with DAPI (4′,6-diamindino-2-phenylindole). Appl Environ Microbiol. 1997. 63:1564–9.24). Lee SJ., Lee YW., Chung J., Lee JK., Lee JY., Jahng D, et al. Reuse of low concentrated electronic wastewater using selected microbe immobilised cell system. Water Sci Technol. 2008. 57:1191–7.
Article25). Bencko V. Use of human hair as a biomarker in the assessment of exposure to pollutants in occupational and environmental settings. Toxicology. 1995. 101:29–39.
Article26). Nakao T., Aozasa O., Ohta S., Miyata H. Assessment of human exposure to PCDDs, PCDFs and Co-PCBs using hair as a human pollution indicator sample I, development of analytical method for human hair and evaluation for exposure assessment. Chemosphere. 2002. 48:885–96.
Article27). Zhang H., Chai Z., Sun H. Human hair as a potential biomonitor for assessing persistent organic pollutants. Environ Int. 2007. 33:685–93.
Article28). Schramm KW. Hair-biomonitoring of organic pollutants. Chemosphere. 2008. 72:1103–11.
Article29). Komaru A., Uchimura Y., Leyama H., Wada KT. Detection of induced triploid scallop, Chlamys nobilis, by DNA microfluorometry with DAPI staining. Aquaculture. 1988. 69:201–9.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Variation and Characterization of Bacterial Communities Contaminating Two Saunas Operated at 64degrees C and 76degrees C
- Characterization of Bacterial Community Contaminating Floor of A Hot and Dry Sauna
- Criteria for Critique of Qualitative Nursing Research
- The Trend of Qualitative Research in Diabetes
- Qualitative Research in Healthcare: Necessity and Characteristics