Yonsei Med J.
2012 Mar;53(2):377-385. 10.3349/ymj.2012.53.2.377.
CpG Array Analysis of Histone H3 Lysine 4 Trimethylation by Chromatin Immunoprecipitation Linked to Microarrays Analysis in Peripheral Blood Mononuclear Cells of IgA Nephropathy Patients
- Affiliations
-
- 1Key Laboratory of Laboratory Medical Diagnostics, Ministry of Education, Chongqing Medical University, Chongqing, China. daiyong22@yahoo.com.cn
- 2Kidney Transplantation and Hemopurification Center of PLA, 181th Hospital of Guangzhou Military Area of PLA, Guangxi, China.
- KMID: 1120206
- DOI: http://doi.org/10.3349/ymj.2012.53.2.377
Abstract
- PURPOSE
The purpose of the present study was to investigate the aberrance of histone H3 lysine 4 trimethylation (H3K4me3) in patients with IgA Nephropathy (IgAN).
MATERIALS AND METHODS
In this study, H3K4me3 variations in peripheral blood mononuclear cells (PBMCs) from 15 IgAN patients and 15 healthy subjects were analyzed using chromatin immunoprecipitation linked to microarrays analysis (ChIP-chip). ChIP real-time PCR was used to validate the microarray results. Expression analysis by quantitative real-time PCR (qRT-PCR) revealed correlations between mRNA and H3K4me3 levels. DNA methylation status was analyzed by quantitative methylation-specific PCR.
RESULTS
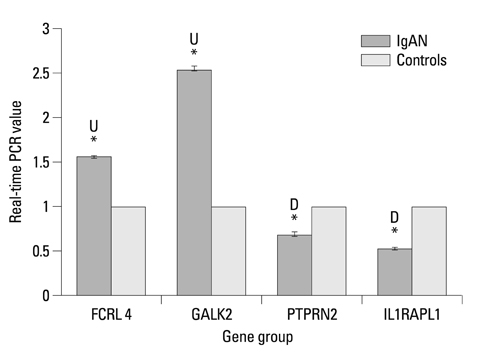
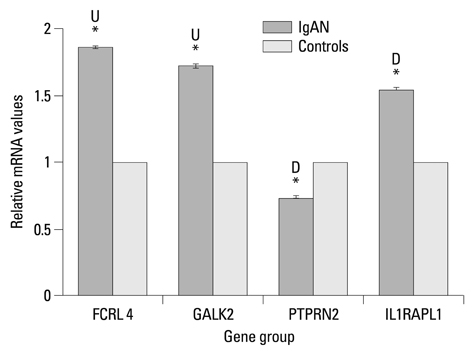
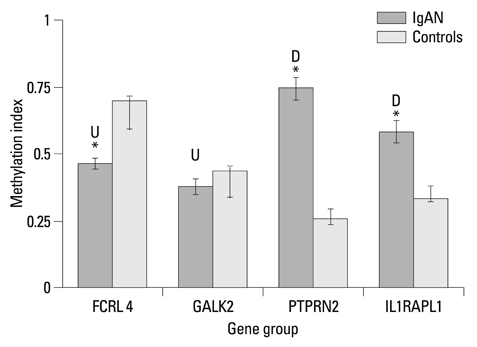
We found that 321 probes displayed significant H3K4me3 differences in IgAN patients compared with healthy controls. Among these probes, 154 probes displayed increased H3K4me3 and 167 probes demonstrated decreased H3K4me3. For further validation, we selected 4 key relevant genes (FCRL4, GALK2, PTPRN2 and IL1RAPL1) to study. The results of ChIP real-time PCR coincided well with the microarray data. Quantitative RT-PCR revealed the correlations between the mRNA expression and the methylation levels of H3K4me3. Different degrees of DNA methylation alterations appeared on the selected positive genes.
CONCLUSION
Our studies indicated that there were significant alterations in H3K4me3 in IgAN patients. These findings may help to explain the disturbed immunity and abnormal glycosylation involved in IgAN patients.
Keyword
MeSH Terms
Figure
Reference
-
1. Eijgenraam JW, van Kooten C. IgA1 glycosylation in IgA nephropathy: as sweet as it can be. Kidney Int. 2008. 73:1106–1108.
Article2. Lai KN. Pathogenic IgA in IgA nephropathy: still the blind men and the elephant? Kidney Int. 2006. 69:1102–1103.
Article3. Julian BA, Novak J. IgA nephropathy: an update. Curr Opin Nephrol Hypertens. 2004. 13:171–179.
Article4. Novak J, Julian BA, Tomana M, Mestecky J. IgA glycosylation and IgA immune complexes in the pathogenesis of IgA nephropathy. Semin Nephrol. 2008. 28:78–87.
Article5. Zhu L, Tang W, Li G, Lv J, Ding J, Yu L, et al. Interaction between variants of two glycosyltransferase genes in IgA nephropathy. Kidney Int. 2009. 76:190–198.
Article6. Narita I, Gejyo F. Pathogenetic significance of aberrant glycosylation of IgA1 in IgA nephropathy. Clin Exp Nephrol. 2008. 12:332–338.
Article7. Rifai A. IgA nephropathy: immune mechanisms beyond IgA mesangial deposition. Kidney Int. 2007. 72:239–241.
Article8. Barratt J, Smith AC, Molyneux K, Feehally J. Immunopathogenesis of IgAN. Semin Immunopathol. 2007. 29:427–443.
Article9. Baylin SB, Herman JG. DNA hypermethylation in tumorigenesis: epigenetics joins genetics. Trends Genet. 2000. 16:168–174.
Article10. Esteller M. CpG island hypermethylation and tumor suppressor genes: a booming present, a brighter future. Oncogene. 2002. 21:5427–5440.
Article11. Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, et al. Active genes are tri-methylated at K4 of histone H3. Nature. 2002. 419:407–411.
Article12. Schneider R, Bannister AJ, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Histone H3 lysine 4 methylation patterns in higher eukaryotic genes. Nat Cell Biol. 2004. 6:73–77.
Article13. Bannister AJ, Kouzarides T. Histone methylation: recognizing the methyl mark. Methods Enzymol. 2004. 376:269–288.
Article14. Bernstein BE, Kamal M, Lindblad-Toh K, Bekiranov S, Bailey DK, Huebert DJ, et al. Genomic maps and comparative analysis of histone modifications in human and mouse. Cell. 2005. 120:169–181.
Article15. Martin C, Zhang Y. The diverse functions of histone lysine methylation. Nat Rev Mol Cell Biol. 2005. 6:838–849.
Article16. Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G, et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet. 2005. 37:391–400.
Article17. Wu J, Smith LT, Plass C, Huang TH. ChIP-chip comes of age for genome-wide functional analysis. Cancer Res. 2006. 66:6899–6902.
Article18. Opel M, Lando D, Bonilla C, Trewick SC, Boukaba A, Walfridsson J, et al. Genome-wide studies of histone demethylation catalysed by the fission yeast homologues of mammalian LSD1. PLoS One. 2007. 2:e386.
Article19. Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, et al. Genome-wide location and function of DNA binding proteins. Science. 2000. 290:2306–2309.
Article20. Weinmann AS, Yan PS, Oberley MJ, Huang TH, Farnham PJ. Isolating human transcription factor targets by coupling chromatin immunoprecipitation and CpG island microarray analysis. Genes Dev. 2002. 16:235–244.
Article21. Miao F, Wu X, Zhang L, Yuan YC, Riggs AD, Natarajan R. Genome-wide analysis of histone lysine methylation variations caused by diabetic conditions in human monocytes. J Biol Chem. 2007. 282:13854–13863.
Article22. Zhang L, Zhong K, Dai Y, Zhou H. Genome-wide analysis of histone H3 lysine 27 trimethylation by ChIP-chip in gastric cancer patients. J Gastroenterol. 2009. 44:305–312.
Article23. Kondo Y, Shen L, Cheng AS, Ahmed S, Boumber Y, Charo C, et al. Gene silencing in cancer by histone H3 lysine 27 trimethylation independent of promoter DNA methylation. Nat Genet. 2008. 40:741–750.
Article24. Haas M. Histologic subclassification of IgA nephropathy: a clinicopathologic study of 244 cases. Am J Kidney Dis. 1997. 29:829–842.
Article25. Lan K, Verma SC, Murakami M, Bajaj B, Robertson ES. Isolation of human peripheral blood mononuclear cells (PBMCs). Curr Protoc Microbiol. 2007. Appendix 4:Appendix 4C.
Article26. Wu H, Ji H. JAMIE: joint analysis of multiple ChIP-chip experiments. Bioinformatics. 2010. 26:1864–1870.
Article27. Kim S, Hu J, Oh Y, Park J, Choi J, Lee YH, et al. Combining ChIP-chip and expression profiling to model the MoCRZ1 mediated circuit for Ca/calcineurin signaling in the rice blast fungus. PLoS Pathog. 2010. 6:e1000909.28. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001. 25:402–408.
Article29. Luszczek W, Cheriyath V, Mekhail TM, Borden EC. Combinations of DNA methyltransferase and histone deacetylase inhibitors induce DNA damage in small cell lung cancer cells: correlation of resistance with IFN-stimulated gene expression. Mol Cancer Ther. 2010. 9:2309–2321.
Article30. Durr ML, Mydlarz WK, Shao C, Zahurak ML, Chuang AY, Hoque MO, et al. Quantitative methylation profiles for multiple tumor suppressor gene promoters in salivary gland tumors. PLoS One. 2010. 5:e10828.
Article31. Liu Q, Gong Z. The coupling of epigenome replication with DNA replication. Curr Opin Plant Biol. 2011. 14:187–194.
Article32. Roh TY, Cuddapah S, Cui K, Zhao K. The genomic landscape of histone modifications in human T cells. Proc Natl Acad Sci U S A. 2006. 103:15782–15787.
Article33. Heisler LE, Torti D, Boutros PC, Watson J, Chan C, Winegarden N, et al. CpG Island microarray probe sequences derived from a physical library are representative of CpG Islands annotated on the human genome. Nucleic Acids Res. 2005. 33:2952–2961.
Article34. Kanamaru Y, Arcos-Fajardo M, Moura IC, Tsuge T, Cohen H, Essig M, et al. Fc alpha receptor I activation induces leukocyte recruitment and promotes aggravation of glomerulonephritis through the FcR gamma adaptor. Eur J Immunol. 2007. 37:1116–1128.
Article35. Ding JX, Xu LX, Zhu L, Lv JC, Zhao MH, Zhang H, et al. Activity of alpha2,6-sialyltransferase and its gene expression in peripheral B lymphocytes in patients with IgA nephropathy. Scand J Immunol. 2009. 69:174–180.
Article36. Thoden JB, Holden HM. The molecular architecture of human N-acetylgalactosamine kinase. J Biol Chem. 2005. 280:32784–32791.
Article37. Fuchs M, Müller T, Lerch MM, Ullrich A. Association of human protein-tyrosine phosphatase kappa with members of the armadillo family. J Biol Chem. 1996. 271:16712–16719.
Article38. Bukowski RM. Tyrosine kinase inhibitors in advanced renal cell carcinoma: the evolving treatment paradigm. Clin Genitourin Cancer. 2009. 7:9–10.
Article39. Navis AC, van den Eijnden M, Schepens JT, Hooft van Huijsduijnen R, Wesseling P, Hendriks WJ. Protein tyrosine phosphatases in glioma biology. Acta Neuropathol. 2010. 119:157–175.
Article40. Guz J, Foksiński M, Oliński R. [Global DNA hipomethylation--the meaning in carcinogenesis]. Postepy Biochem. 2010. 56:16–21.41. Hahn WH, Cho BS, Kim SD, Kim SK, Kang S. Interleukin-1 cluster gene polymorphisms in childhood IgA nephropathy. Pediatr Nephrol. 2009. 24:1329–1336.
Article42. Maunakea AK, Nagarajan RP, Bilenky M, Ballinger TJ, D'Souza C, Fouse SD, et al. Conserved role of intragenic DNA methylation in regulating alternative promoters. Nature. 2010. 466:253–257.
Article43. Kim DS, Kim MJ, Lee JY, Kim YZ, Kim EJ, Park JY. Aberrant methylation of E-cadherin and H-cadherin genes in nonsmall cell lung cancer and its relation to clinicopathologic features. Cancer. 2007. 110:2785–2792.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Histone lysine demethylases in mammalian embryonic development
- Coordinated transcriptional regulation of calmegin, a testis-specific molecular chaperon, by histone deacetylase and CpG methyltransferase
- Role of mononuclear cells of IgA nephropathy on ICAM-1 expression in mesangial cells
- The Global Histone Modification Patterns of Osteosarcoma
- A Review of Three Different Studies on Hidden Markov Models for Epigenetic Problems: A Computational Perspective