Yonsei Med J.
2008 Apr;49(2):301-310. 10.3349/ymj.2008.49.2.301.
Identification of Up-Regulated Genes in Malignant Glioma with Subtraction Hybridization: Preliminary Screening Studies
- Affiliations
-
- 1Department of Neurosurgery, Ewha Womans University, School of Medicine, Seoul, Korea. yongcho@ewha.ac.kr
- KMID: 1084505
- DOI: http://doi.org/10.3349/ymj.2008.49.2.301
Abstract
- PURPOSE
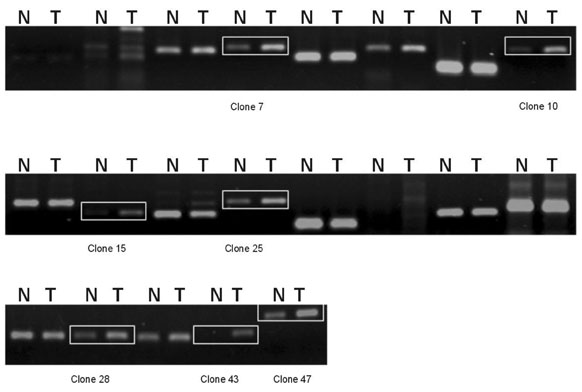
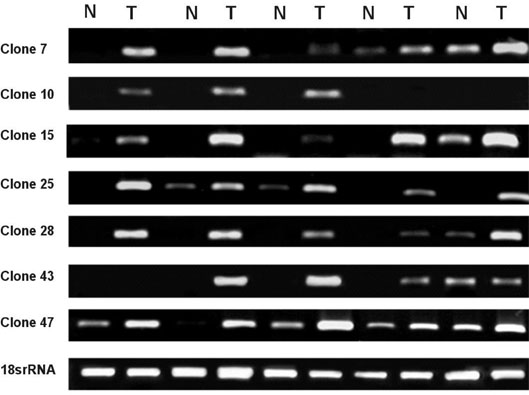
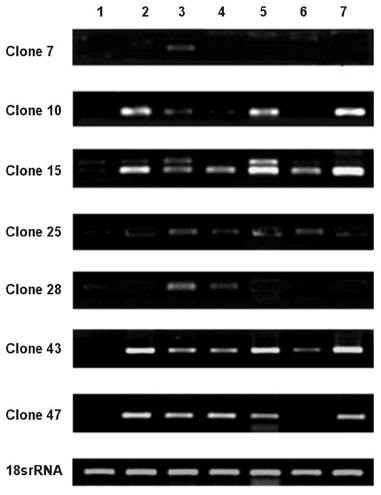
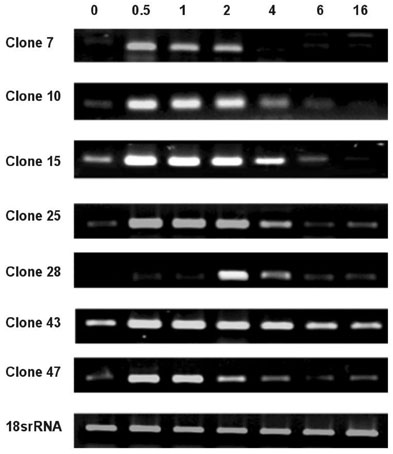
This investigation is intended to obtain differentially expressed genes related to human malignant glioma using Subtractive hybridization. MATERIALS AND METHODS: Subtractive hybridization is potentially faster methods for identifying differentially expressed genes associated with a particular disease state. We identified 7 over-expressed genes which were not homologous to any of the known genes in the Genbank(TM) database. RESULTS: Using semi-quantitative reverse transcription-polymerase chain reaction (RT-PCR), the mRNA expression levels of these 7 genes were higher in human glioblastomas tissue than in non-tumor brain tissue. In order to learn more about the expression profile of these genes, RT-PCR was performed using various commercially available human carcinoma cell lines. Some of these new genes were over-expressed in human glioma cell line, but not the expressed in other human cancer cell line. CONCLUSION: Theses cloned new genes may play a role in brain tumorigenesis. Further studies including verification of oncogene, cancer protein, and glioblastoma induction in animal model are needed.
Keyword
MeSH Terms
Figure
Reference
-
1. Dai C, Holland EC. Astrocyte differentiation states and gliomas formation. Cancer J. 2003. 9:72–81.2. Sallinen SL, Sallinen PK, Haapasalo HK, Helin HJ, Helén PT, Schraml P, et al. Identification of differentially expressed genes in human gliomas by DNA microarray and tissue chip techniques. Cancer Res. 2000. 60:6617–6622.3. Christensen HC, Kosteljanetz M, Johansen C. Incidences of gliomas and meningiomas in Denmark, 1943 to 1997. Neurosurgery. 2003. 52:1327–1333. discussion 1333-4.
Article4. Velculescu VE, Zhang L, Zhou W, Vogelstein J, Basrai MA, Bassett DE Jr, et al. Characterization of the yeast transcriptome. Cell. 1997. 88:243–251.
Article5. Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001. 11:703–715.
Article6. Nutt CL, Zerillo CA, Kelly GM, Hockfield S. Brain enriched hyaluronan binding (BEHAB)/brevican increases aggressiveness of CNS-1 gliomas in Lewis rats. Cancer Res. 2001. 61:7056–7059.7. Palackal NT, Lee SH, Harvey RG, Blair IA, Penning TM. Activation of polycyclic aromatic hydrocarbon trans-dihydrodiol proximate carcinogens by human aldo-keto reductase (AKR1C) enzymes and their functional overexpression in human lung carcinoma (A549) cells. J Biol Chem. 2002. 277:24799–24808.
Article8. Rehman I, Cross SS, Azzouzi AR, Catto JW, Deloulme JC, Larre S, et al. S100A6 (Calcyclin) is a prostate basal cell marker absent in prostate cancer and its precursors. Br J Cancer. 2004. 91:739–744.
Article9. Welsh J, Chada K, Dalal SS, Cheng R, Ralph D, McClelland M. Arbitrarily primed PCR fingerprinting of RNA. Nucleic Acids Res. 1992. 20:4965–4970.
Article10. Smith JM, Bowles J, Wilson M, Koopman P. HMG box transcription factor gene Hbp1 is expressed in germ cells of the developing mouse testis. Dev Dyn. 2004. 230:366–370.
Article11. van Nocker S, Ludwig P. The WD-repeat protein superfamily in Arabidopsis: conservation and divergence in structure and function. BMC Genomics. 2003. 4:50.
Article12. Porkka KP, Visakorpi T. Detection of differentially expressed genes in prostate cancer by combining suppression subtractive hybridization and cDNA library array. J Pathol. 2001. 193:73–79.
Article13. Wandrey M, Trevaskis B, Brewin N, Udvardi MK. Molecular and cell biology of a family of voltagedependent anion channel porins in Lotus japonicus. Plant Physiol. 2004. 134:182–193.
Article14. Zeindl-Eberhart E, Haraida S, Liebmann S, Jungblut PR, Lamer S, Mayer Mayer, et al. Detection and identification of tumor-associated protein variants in human hepatocellular carcinomas. Hepatology. 2004. 39:540–549.
Article15. Diatchenko L, Lau YF, Campbell AP, Chenchik A, Moqadam F, Huang B, et al. Suppression subtractive hybridization: a method for generating differentially regulated or tissue-specific cDNA probes and libraries. Proc Natl Acad Sci U S A. 1996. 93:6025–6030.
Article16. Hufton SE, Moerkerk PT, Brandwijk R, de Bruïne AP, Arends JW, Hoogenboom HR. A profile of differentially expressed genes in primary colorectal cancer using suppression subtractive hybridization. FEBS Lett. 1999. 463:77–82.
Article17. Ji W, Wright MB, Cai L, Flament A, Lindpaintner K. Efficacy of SSH PCR in isolating differentially expressed genes. BMC Genomics. 2002. 3:12.
Article18. Lee SJ, Suh MC, Kim S, Kwon JK, Kim M, Paek KH, et al. Molecular cloning of a novel pathogen-inducible cDNA encoding a putative acyl-CoA synthetase from Capsicum annuum L. Plant Mol Biol. 2001. 46:661–671.19. Shridhar V, Sen A, Chien J, Staub J, Avula R, Kovats S, et al. Identification of underexpressed genes in earlyand late-stage primary ovarian tumors by suppression subtraction hybridization. Cancer Res. 2002. 62:262–270.20. Swearingen ML, Sun D, Bourner M, Weinstein EJ. Detection of differentially expressed HES-6 gene in metastatic colon carcinoma by combination of suppression subtractive hybridization and cDNA library array. Cancer Lett. 2003. 198:229–239.
Article21. Liang P, Pardee AB. Differential display of eukaryotic messenger RNA by means of the polymerase chain reaction. Science. 1992. 257:967–971.
Article22. Dong Y, Zhang H, Hawthorn L, Ganther HE, Ip C. Delineation of the molecular basis for seleniuminduced growth arrest in human prostate cancer cells by oligonucleotide array. Cancer Res. 2003. 63:52–59.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Current Immunotherapeutic Approaches for Malignant Gliomas
- Expression of Preadipocyte Factor-1 (Pref-1) and Vitamin D3 Up-regulated Protein 1 (VDUP1) Genes in Rat Adrenal Gland following Chronic Immobilization Stress
- Molecular Cloning of Novel Genes Related to the Craniofacial Development of Human Embryo
- Next generation sequencing and array-based comparative genomic hybridization for molecular diagnosis of pediatric endocrine disorders
- Fluoxetine Up-Regulates Bcl-xL Expression in Rat C6 Glioma Cells