Cancer Res Treat.
2023 Jul;55(3):969-977. 10.4143/crt.2022.1557.
Early Plasma Circulating Tumor DNA as a Potential Biomarker of Disease Recurrence in Non-metastatic Prostate Cancer
- Affiliations
-
- 1Department of Urology, Ren Ji Hospital, Shanghai Jiao Tong University School of Medicine, Shanghai, China
- KMID: 2544177
- DOI: http://doi.org/10.4143/crt.2022.1557
Abstract
- Purpose
In non-metastatic prostate cancer (nmPCa) setting, it is important to early identify the patients at risk of biochemical recurrence (BCR) for immediate postoperative intervention. Our study aimed to evaluate the potential clinical utility of circulating tumor DNA (ctDNA) for predicting disease recurrence.
Materials and Methods
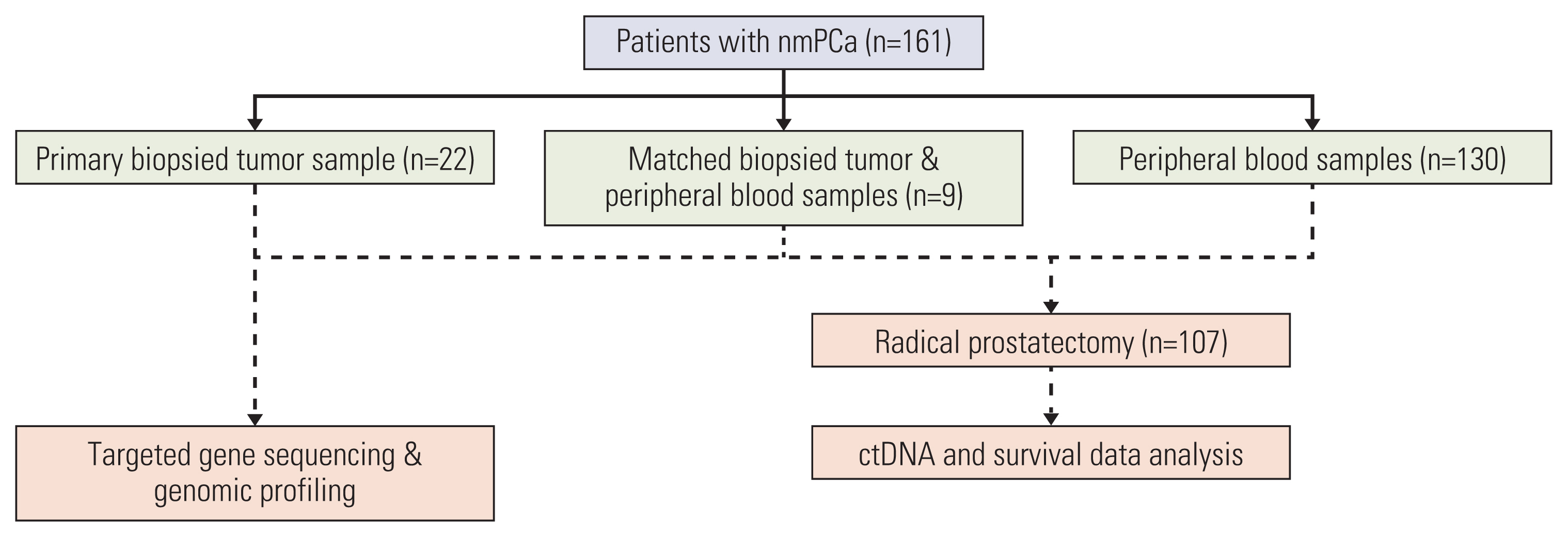
This real-world observational study evaluated 161 cases of nmPCa undergoing next-generation sequencing at our institution. A total of 139 ctDNA samples and 31 biopsied tumor tissue underwent genomic profiling. The study endpoint was BCR after radical prostatectomy. Relationships between the ctDNA status and the biochemical progression-free survival (bPFS) were analyzed by log-rank test and multivariate Cox regression.
Results
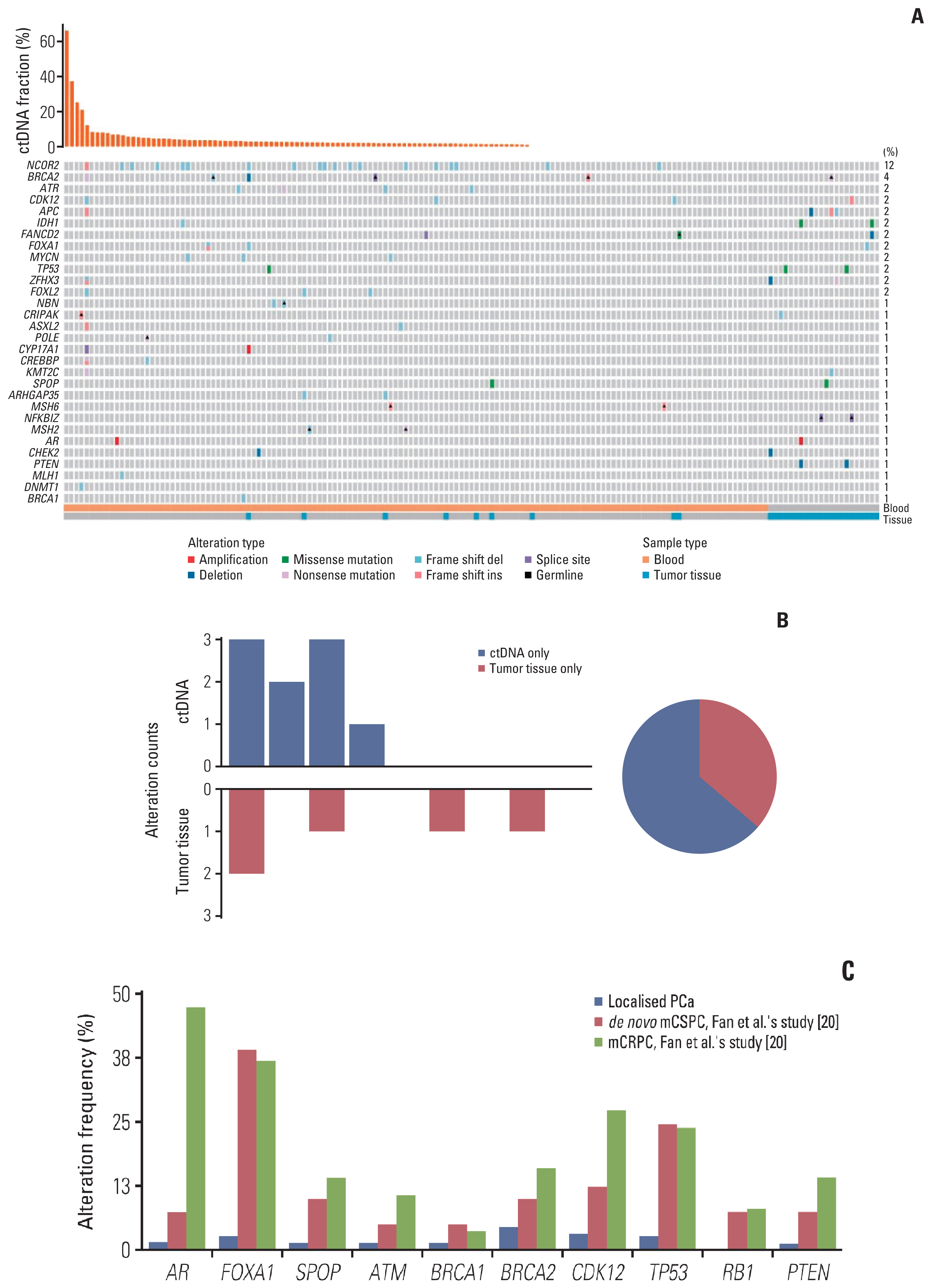
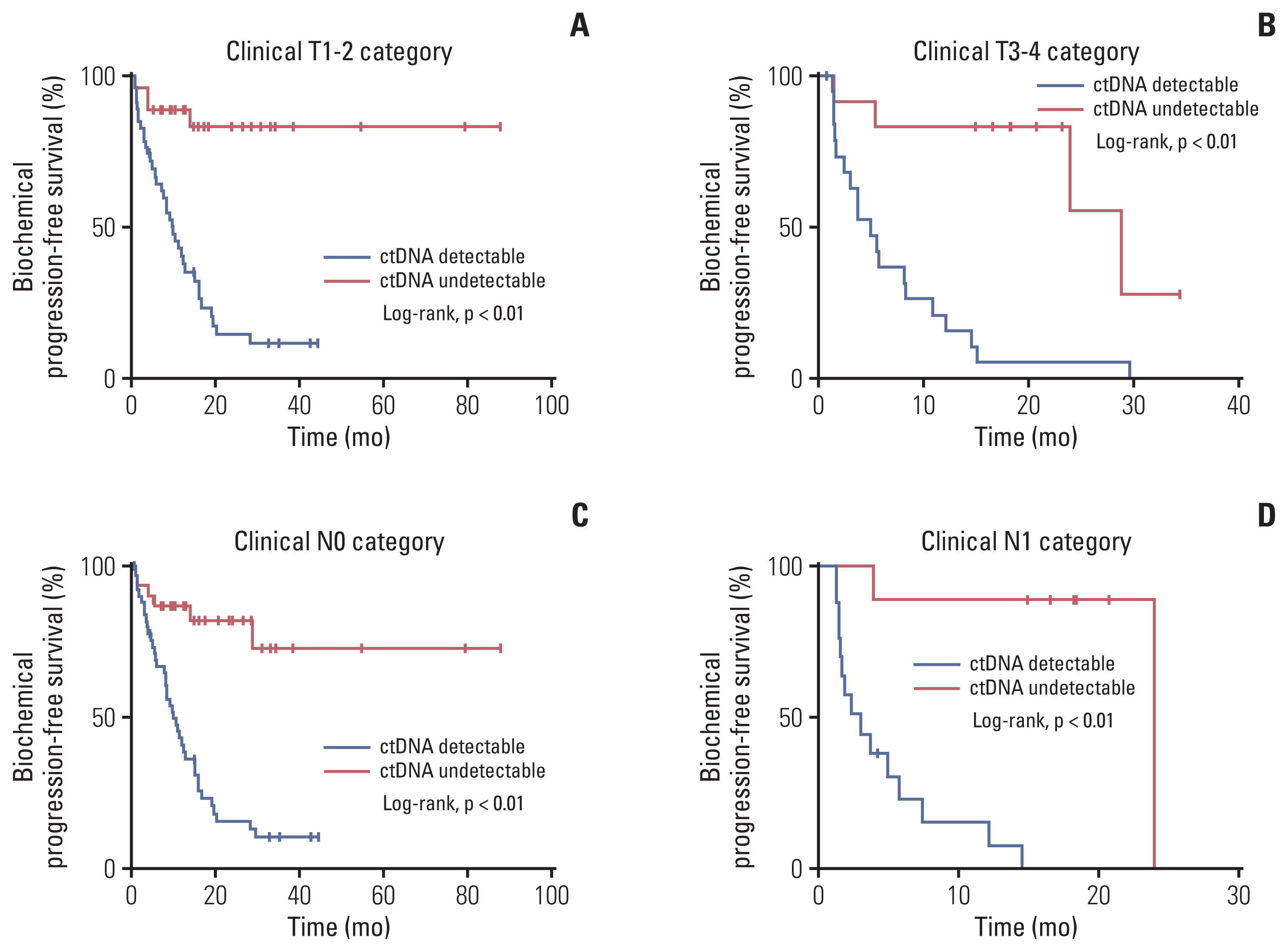
Of 161 enrolled patients, 19 (11.8%) harbored deleterious alterations in NCOR2, followed by BRCA2 (3.7%), ATR (2.5%), and CDK12 (2.5%). Of available pre-operative blood samples (n=139), ctDNA was detectable in 91 (65.5%). Until last follow-up, 56 of 68 patients (85.3%) with detectable ctDNA had achieved BCR, whereas only eight of 39 patients (20.5%) with undetectable ctDNA had achieved BCR. Patients who had undetectable ctDNA experienced significantly longer bPFS compared with those who had detectable ctDNA (not available vs. 8.2 months; hazard ratio, 0.14; p < 0.01). Pre-operative ctDNA status was a significant prognostic factor of disease recurrence.
Conclusion
Pre-operative ctDNA detection could identify patients at high risk of recurrence and has the potential to inform immediate postoperative interventions, but these approaches remain to be validated in prospective studies. ctDNA studies can provide insights into accurate monitoring and precise treatment rather than simply following routine clinical care.
Figure
Reference
-
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020; 70:7–30.
Article2. Zhu Y, Mo M, Wei Y, Wu J, Pan J, Freedland SJ, et al. Epidemiology and genomics of prostate cancer in Asian men. Nat Rev Urol. 2021; 18:282–301.
Article3. Allemani C, Matsuda T, Di Carlo V, Harewood R, Matz M, Niksic M, et al. Global surveillance of trends in cancer survival 2000–14 (CONCORD-3): analysis of individual records for 37 513 025 patients diagnosed with one of 18 cancers from 322 population-based registries in 71 countries. Lancet. 2018; 391:1023–75.4. Mottet N, Bellmunt J, Bolla M, Briers E, Cumberbatch MG, De Santis M, et al. EAU-ESTRO-SIOG guidelines on prostate cancer. Part 1: screening, diagnosis, and local treatment with curative intent. Eur Urol. 2017; 71:618–29.
Article5. van der Toom EE, Axelrod HD, de la Rosette JJ, de Reijke TM, Pienta KJ, Valkenburg KC. Prostate-specific markers to identify rare prostate cancer cells in liquid biopsies. Nat Rev Urol. 2019; 16:7–22.
Article6. Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003; 169:517–23.
Article7. Diehl F, Schmidt K, Choti MA, Romans K, Goodman S, Li M, et al. Circulating mutant DNA to assess tumor dynamics. Nat Med. 2008; 14:985–90.
Article8. Vandekerkhove G, Struss WJ, Annala M, Kallio HM, Khalaf D, Warner EW, et al. Circulating tumor DNA abundance and potential utility in de novo metastatic prostate cancer. Eur Urol. 2019; 75:667–75.
Article9. Bettegowda C, Sausen M, Leary RJ, Kinde I, Wang Y, Agrawal N, et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci Transl Med. 2014; 6:224ra24.10. Lee B, Lipton L, Cohen J, Tie J, Javed AA, Li L, et al. Circulating tumor DNA as a potential marker of adjuvant chemotherapy benefit following surgery for localized pancreatic cancer. Ann Oncol. 2019; 30:1472–8.
Article11. Magbanua MJM, Swigart LB, Wu HT, Hirst GL, Yau C, Wolf DM, et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann Oncol. 2021; 32:229–39.12. Sumanasuriya S, Seed G, Parr H, Christova R, Pope L, Bertan C, et al. Elucidating prostate cancer behaviour during treatment via low-pass whole-genome sequencing of circulating tumour DNA. Eur Urol. 2021; 80:243–53.
Article13. Raja R, Kuziora M, Brohawn PZ, Higgs BW, Gupta A, Dennis PA, et al. Early reduction in ctDNA predicts survival in patients with lung and bladder cancer treated with durvalumab. Clin Cancer Res. 2018; 24:6212–22.
Article14. Yang J, Gong Y, Lam VK, Shi Y, Guan Y, Zhang Y, et al. Deep sequencing of circulating tumor DNA detects molecular residual disease and predicts recurrence in gastric cancer. Cell Death Dis. 2020; 11:346.
Article15. Tie J, Cohen JD, Wang Y, Li L, Christie M, Simons K, et al. Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut. 2019; 68:663–71.
Article16. Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med. 2016; 8:346ra92.
Article17. Garcia-Murillas I, Schiavon G, Weigelt B, Ng C, Hrebien S, Cutts RJ, et al. Mutation tracking in circulating tumor DNA predicts relapse in early breast cancer. Sci Transl Med. 2015; 7:302ra133.
Article18. Dong B, Fan L, Yang B, Chen W, Li Y, Wu K, et al. Use of circulating tumor DNA for the clinical management of metastatic castration-resistant prostate cancer: a multicenter, real-world study. J Natl Compr Canc Netw. 2021; 19:905–14.
Article19. Vandekerkhove G, Todenhofer T, Annala M, Struss WJ, Wong A, Beja K, et al. Circulating tumor DNA reveals clinically actionable somatic genome of metastatic bladder cancer. Clin Cancer Res. 2017; 23:6487–97.
Article20. Fan L, Fei X, Zhu Y, Pan J, Sha J, Chi C, et al. Comparative analysis of genomic alterations across castration sensitive and castration resistant prostate cancer via circulating tumor DNA sequencing. J Urol. 2021; 205:461–9.
Article21. Cancer Genome Atlas Research Network. The molecular taxonomy of primary prostate cancer. Cell. 2015; 163:1011–25.22. Hennigan ST, Trostel SY, Terrigino NT, Voznesensky OS, Schaefer RJ, Whitlock NC, et al. Low abundance of circulating tumor DNA in localized prostate cancer. JCO Precis Oncol. 2019; 3:PO.1900176.
Article23. Chen E, Cario CL, Leong L, Lopez K, Marquez CP, Li PS, et al. Cell-free DNA detection of tumor mutations in heterogeneous, localized prostate cancer via targeted, multiregion sequencing. JCO Precis Oncol. 2021; 5:PO20.00428.
Article24. Li J, Xu C, Lee HJ, Ren S, Zi X, Zhang Z, et al. A genomic and epigenomic atlas of prostate cancer in Asian populations. Nature. 2020; 580:93–9.25. Fraser M, Sabelnykova VY, Yamaguchi TN, Heisler LE, Livingstone J, Huang V, et al. Genomic hallmarks of localized, non-indolent prostate cancer. Nature. 2017; 541:359–64.26. Jamaspishvili T, Berman DM, Ross AE, Scher HI, De Marzo AM, Squire JA, et al. Clinical implications of PTEN loss in prostate cancer. Nat Rev Urol. 2018; 15:222–34.
Article27. Lau E, McCoy P, Reeves F, Chow K, Clarkson M, Kwan EM, et al. Detection of ctDNA in plasma of patients with clinically localised prostate cancer is associated with rapid disease progression. Genome Med. 2020; 12:72.28. Chen K, Zhao H, Shi Y, Yang F, Wang LT, Kang G, et al. Perioperative dynamic changes in circulating tumor DNA in patients with lung cancer (DYNAMIC). Clin Cancer Res. 2019; 25:7058–67.29. Tan L, Sandhu S, Lee RJ, Li J, Callahan J, Ftouni S, et al. Prediction and monitoring of relapse in stage III melanoma using circulating tumor DNA. Ann Oncol. 2019; 30:804–14.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Application of Circulating Tumor DNA Analysis
- Identification of Enhancer of Zeste Homolog 2 Expression in Peripheral Circulating Tumor Cells in Metastatic Prostate Cancer Patients: A Preliminary Study
- Circulating Tumor Cell and Cell-free Circulating Tumor DNA in Lung Cancer
- Evaluation of Circulating Tumor DNA in Patients with Ovarian Cancer Harboring Somatic PIK3CA or KRAS Mutations
- Selective utilization of circulating tumor DNA testing enables disease monitoring in endometrial and ovarian carcinomas