Cancer Res Treat.
2023 Jul;55(3):927-938. 10.4143/crt.2023.268.
Circulating Tumor DNA Dynamics and Treatment Outcome of Regorafenib in Metastatic Colorectal Cancer
- Affiliations
-
- 1Department of Internal Medicine, Seoul National University Hospital, Seoul, Korea
- 2IMBdx, Inc., Seoul, Korea
- 3Cancer Research Institute, Seoul National University College of Medicine, Seoul, Korea
- 4Department of Surgery, Seoul National University Hospital, Seoul, Korea
- 5Department of Pathology, Seoul National University College of Medicine, Seoul, Korea
- 6Department of Molecular Medicine and Biopharmaceutical Sciences, Graduate School of Convergence Science and Technology, Seoul National University, Seoul, Korea
- KMID: 2544173
- DOI: http://doi.org/10.4143/crt.2023.268
Abstract
- Purpose
Circulating tumor DNA (ctDNA) is emerging as a valuable non-invasive tool to identify tumor heterogeneity and tumor burden. This study investigated ctDNA dynamics in metastatic colorectal cancer patients treated with regorafenib.
Materials and Methods
In this prospective biomarker study, plasma cell-free DNA (cfDNA) samples obtained at baseline, at the first response evaluation after 2 cycles of treatment, and at the time of progressive disease were sequenced using a targeted next-generation sequencing platform which included 106 genes.
Results
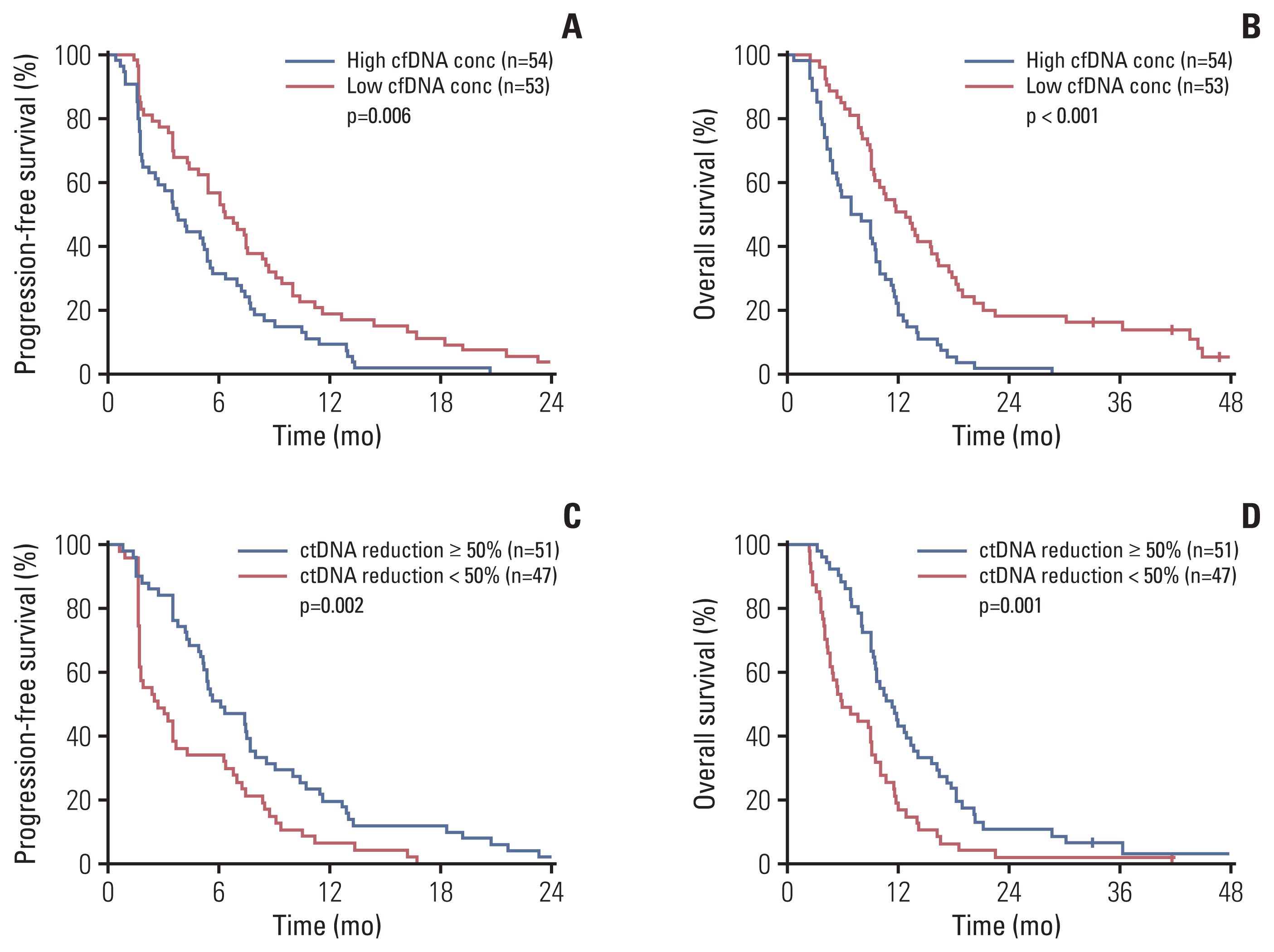
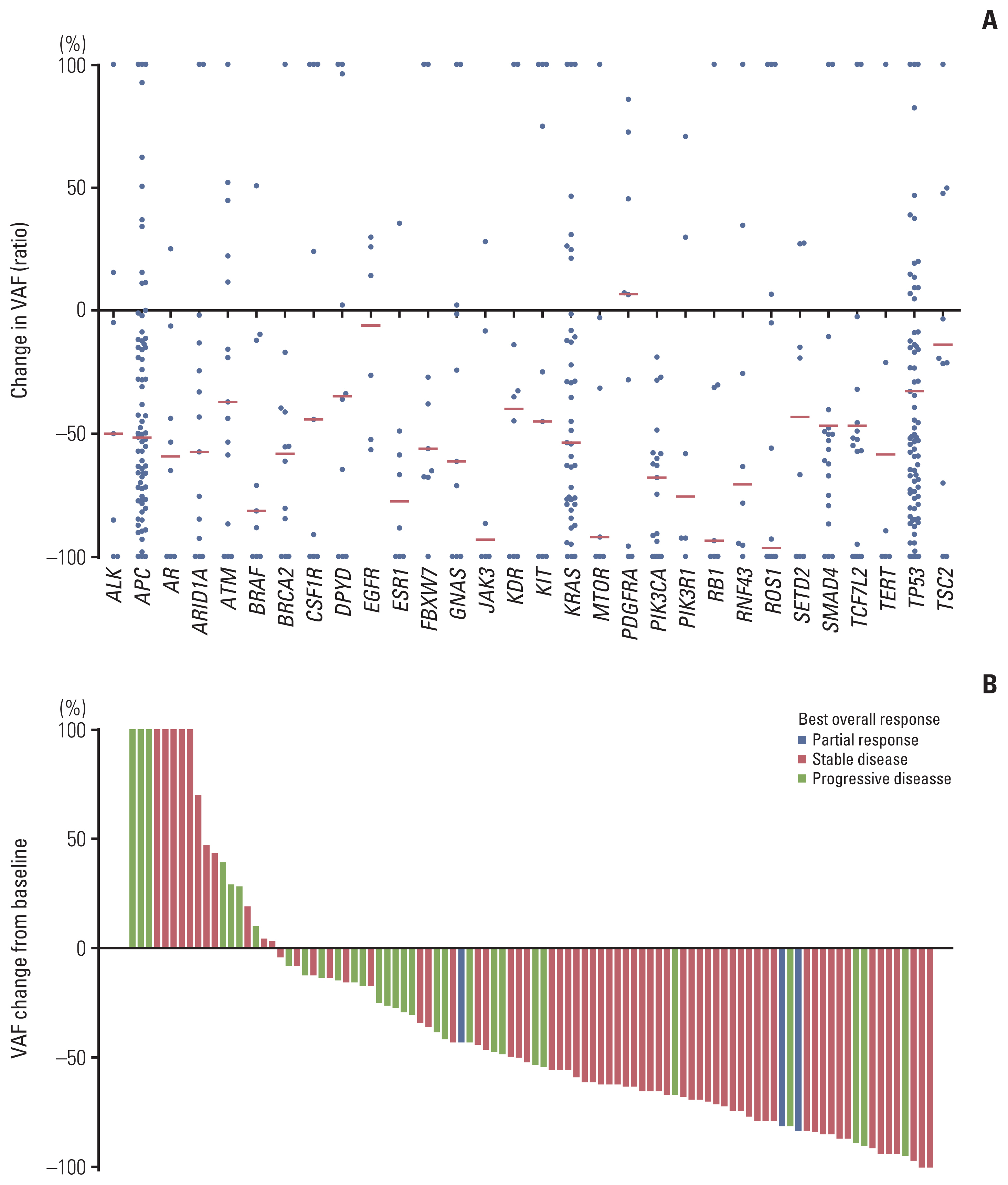
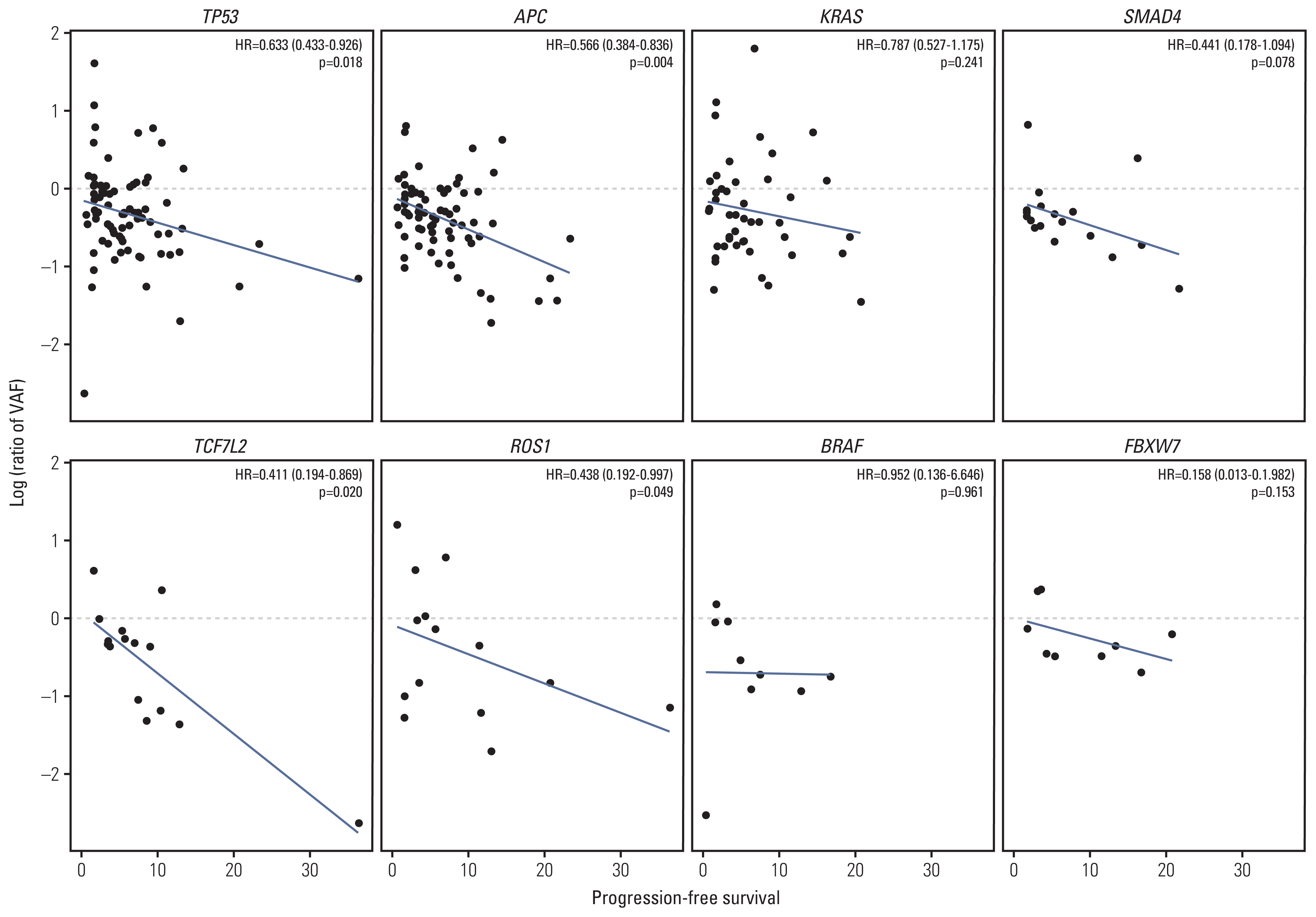
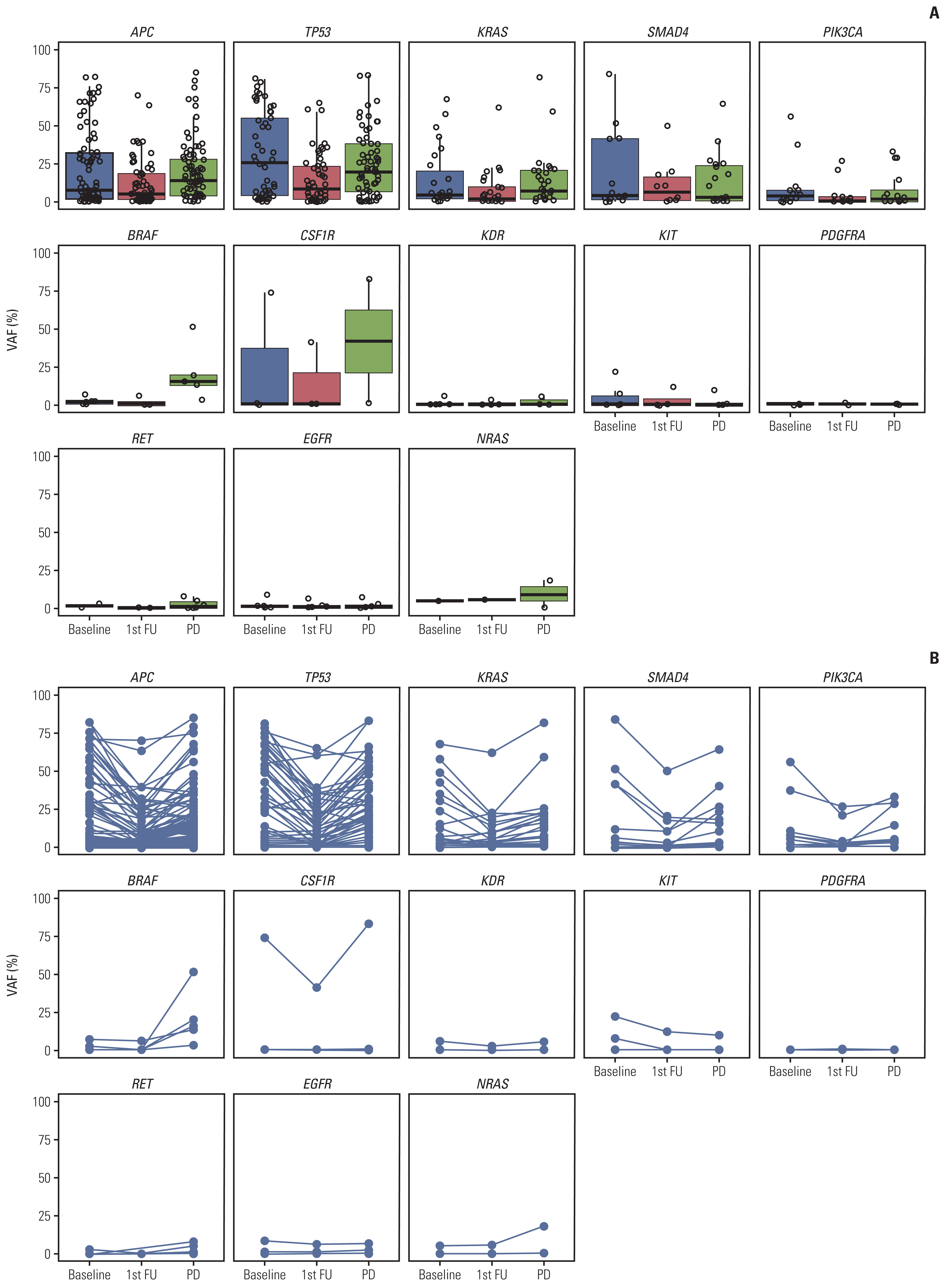
A total of 285 blood samples from 110 patients were analyzed. Higher baseline cfDNA concentration was associated with worse progression-free survival (PFS) and overall survival (OS). After 2 cycles of treatment, variant allele frequency (VAF) in the majority of ctDNA mutations decreased with a mean relative change of –31.6%. Decreases in the VAF of TP53, APC, TCF7L2, and ROS1 after 2 cycles of regorafenib were associated with longer PFS. We used the sum of VAF at each time point as a surrogate for the overall ctDNA burden. A reduction in sum (VAF) of ≥ 50% after 2 cycles was associated with longer PFS (6.1 vs. 2.7 months, p=0.002), OS (11.3 vs. 5.9 months, p=0.001), and higher disease control rate (86.3% vs. 51.1%, p < 0.001). VAF of the majority of the ctDNA mutations increased at the time of disease progression, and VAF of BRAF increased markedly.
Conclusion
Reduction in ctDNA burden as estimated by sum (VAF) could be used to predict treatment outcome of regorafenib.
Keyword
Figure
Reference
-
References
1. Wilhelm SM, Dumas J, Adnane L, Lynch M, Carter CA, Schutz G, et al. Regorafenib (BAY 73–4506): a new oral multikinase inhibitor of angiogenic, stromal and oncogenic receptor tyrosine kinases with potent preclinical antitumor activity. Int J Cancer. 2011; 129:245–55.
Article2. Abou-Elkacem L, Arns S, Brix G, Gremse F, Zopf D, Kiessling F, et al. Regorafenib inhibits growth, angiogenesis, and metastasis in a highly aggressive, orthotopic colon cancer model. Mol Cancer Ther. 2013; 12:1322–31.3. Grothey A, Van Cutsem E, Sobrero A, Siena S, Falcone A, Ychou M, et al. Regorafenib monotherapy for previously treated metastatic colorectal cancer (CORRECT): an international, multicentre, randomised, placebo-controlled, phase 3 trial. Lancet. 2013; 381:303–12.
Article4. Li J, Qin S, Xu R, Yau TC, Ma B, Pan H, et al. Regorafenib plus best supportive care versus placebo plus best supportive care in Asian patients with previously treated metastatic colorectal cancer (CONCUR): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2015; 16:619–29.
Article5. Tabernero J, Lenz HJ, Siena S, Sobrero A, Falcone A, Ychou M, et al. Analysis of circulating DNA and protein biomarkers to predict the clinical activity of regorafenib and assess prognosis in patients with metastatic colorectal cancer: a retrospective, exploratory analysis of the CORRECT trial. Lancet Oncol. 2015; 16:937–48.
Article6. Corcoran RB, Chabner BA. Application of Cell-free DNA Analysis to Cancer Treatment. N Engl J Med. 2018; 379:1754–65.
Article7. Dasari A, Morris VK, Allegra CJ, Atreya C, Benson AB 3rd, Boland P, et al. ctDNA applications and integration in colorectal cancer: an NCI Colon and Rectal-Anal Task Forces whitepaper. Nat Rev Clin Oncol. 2020; 17:757–70.
Article8. Tie J, Kinde I, Wang Y, Wong HL, Roebert J, Christie M, et al. Circulating tumor DNA as an early marker of therapeutic response in patients with metastatic colorectal cancer. Ann Oncol. 2015; 26:1715–22.9. Garlan F, Laurent-Puig P, Sefrioui D, Siauve N, Didelot A, Sarafan-Vasseur N, et al. Early evaluation of circulating tumor DNA as marker of therapeutic efficacy in metastatic colorectal cancer patients (PLACOL Study). Clin Cancer Res. 2017; 23:5416–25.
Article10. Lai Z, Markovets A, Ahdesmaki M, Chapman B, Hofmann O, McEwen R, et al. VarDict: a novel and versatile variant caller for next-generation sequencing in cancer research. Nucleic Acids Res. 2016; 44:e108.
Article11. Cingolani P, Platts A, Wang LL, Coon M, Nguyen T, Wang L, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin). 2012; 6:80–92.
Article12. Cingolani P, Patel VM, Coon M, Nguyen T, Land SJ, Ruden DM, et al. Using Drosophila melanogaster as a model for genotoxic chemical mutational studies with a new program, SnpSift. Front Genet. 2012; 3:35.
Article13. McLaren W, Gil L, Hunt SE, Riat HS, Ritchie GR, Thormann A, et al. The Ensembl variant effect predictor. Genome Biol. 2016; 17:122.
Article14. Strijker M, Soer EC, de Pastena M, Creemers A, Balduzzi A, Beagan JJ, et al. Circulating tumor DNA quantity is related to tumor volume and both predict survival in metastatic pancreatic ductal adenocarcinoma. Int J Cancer. 2020; 146:1445–56.
Article15. Smith JT, Balar A, Lakhani DA, Kluwe C, Zhao Z, Kopparapu P, et al. Circulating tumor DNA as a biomarker of radiographic tumor burden in SCLC. JTO Clin Res Rep. 2021; 2:100110.
Article16. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012; 487:330–7.17. Van Cutsem E, Cervantes A, Adam R, Sobrero A, Van Krieken JH, Aderka D, et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann Oncol. 2016; 27:1386–422.
Article18. Siravegna G, Bardelli A. Genotyping cell-free tumor DNA in the blood to detect residual disease and drug resistance. Genome Biol. 2014; 15:449.19. Diaz LA Jr, Bardelli A. Liquid biopsies: genotyping circulating tumor DNA. J Clin Oncol. 2014; 32:579–86.20. Montagut C, Dalmases A, Bellosillo B, Crespo M, Pairet S, Iglesias M, et al. Identification of a mutation in the extracellular domain of the epidermal growth factor receptor conferring cetuximab resistance in colorectal cancer. Nat Med. 2012; 18:221–3.
Article21. Parseghian CM, Napolitano S, Loree JM, Kopetz S. Mechanisms of innate and acquired resistance to anti-EGFR therapy: a review of current knowledge with a focus on rechallenge therapies. Clin Cancer Res. 2019; 25:6899–908.22. Sartore-Bianchi A, Pietrantonio F, Lonardi S, Mussolin B, Rua F, Fenocchio E, et al. Phase II study of anti-EGFR rechallenge therapy with panitumumab driven by circulating tumor DNA molecular selection in metastatic colorectal cancer: the CHRONOS trial. J Clin Oncol. 2021; 39(15 Suppl):3506.
Article23. Wong AL, Lim JS, Sinha A, Gopinathan A, Lim R, Tan CS, et al. Tumour pharmacodynamics and circulating cell free DNA in patients with refractory colorectal carcinoma treated with regorafenib. J Transl Med. 2015; 13:57.
Article24. Kim S, Lim Y, Kang JK, Kim HP, Roh H, Kim SY, et al. Dynamic changes in longitudinal circulating tumour DNA profile during metastatic colorectal cancer treatment. Br J Cancer. 2022; 127:898–907.
Article25. Van Cutsem E, Martinelli E, Cascinu S, Sobrero A, Banzi M, Seitz JF, et al. Regorafenib for patients with metastatic colorectal cancer who progressed after standard therapy: results of the large, single-arm, open-label phase IIIb CONSIGN study. Oncologist. 2019; 24:185–92.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Clinical Application of Circulating Tumor DNA Analysis
- Current Status of Chemotherapy in Colorectal Cancer: Updated Treatment Strategies
- Role of Liquid Biopsies in Colorectal Cancer
- Clinical Significance of Circulating Tumor Cells in Gastric Cancer
- Circulating Tumor Cell and Cell-free Circulating Tumor DNA in Lung Cancer