J Korean Med Sci.
2023 May;38(19):e145. 10.3346/jkms.2023.38.e145.
Comparison of Newly Proposed LDL-Cholesterol Estimation Equations
- Affiliations
-
- 1Department of Biostatistics, Graduate School, Yonsei University, Seoul, Korea
- 2Yonsei University Wonju Industry-Academic Cooperation Foundation, Wonju, Korea
- 3Division of Endocrinology and Metabolism, Department of Internal Medicine, Hallym University Sacred Heart Hospital, Anyang, Korea
- 4Seoul Clinical Laboratories Biobank, Yongin, Korea
- 5Department of Biomedical Laboratory Science, Songho University, Hoengseong, Korea
- 6Department of Precision Medicine, Wonju College of Medicine, Yonsei University, Wonju, Korea
- KMID: 2542395
- DOI: http://doi.org/10.3346/jkms.2023.38.e145
Abstract
- Background
Low-density lipoprotein cholesterol is an important marker highly associated with cardiovascular disease. Since the direct measurement of it is inefficient in terms of cost and time, it is common to estimate through the Friedewald equation developed about 50 years ago. However, various limitations exist since the Friedewald equation was not designed for Koreans. This study proposes a new low-density lipoprotein cholesterol estimation equation for South Koreans using nationally approved statistical data.
Methods
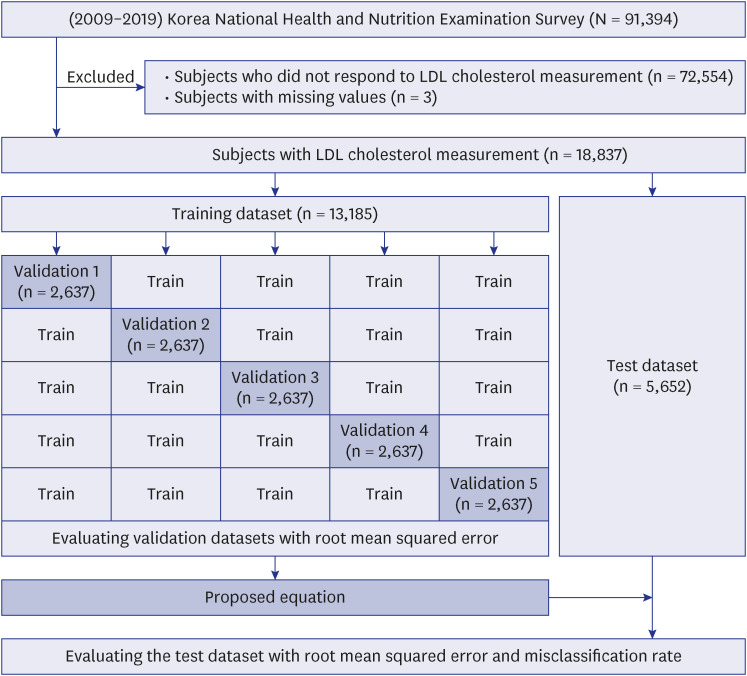
This study used data from the Korean National Health and Nutrition Examination Survey from 2009 to 2019. The 18,837 subjects were used to develop the equation for estimating low-density lipoprotein cholesterol. The subjects included individuals with low-density lipoprotein cholesterol levels directly measured among those with high-density lipoprotein cholesterol, triglycerides, and total cholesterol measured. We compared twelve equations developed in the previous studies and the newly proposed equation (model 1) developed in this study with the actual low-density lipoprotein cholesterol value in various ways.
Results
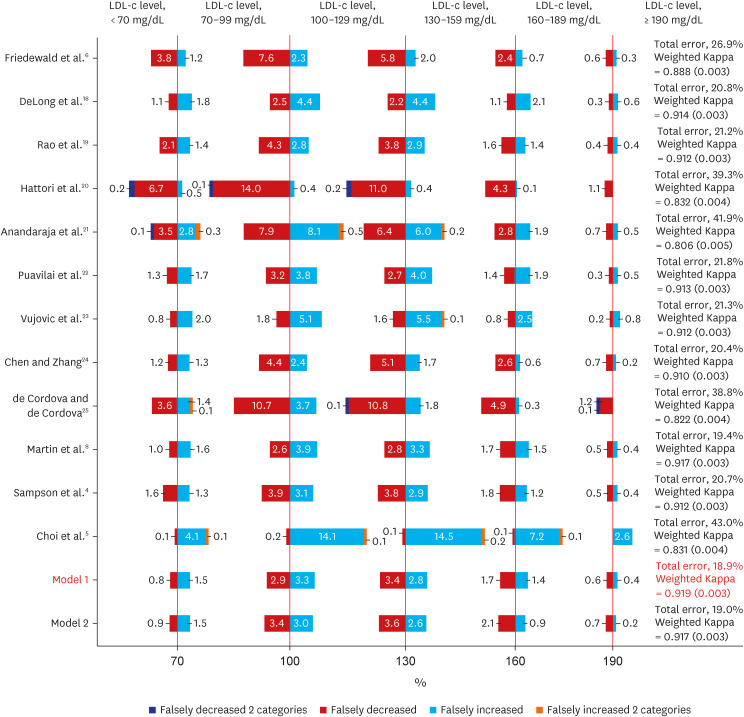
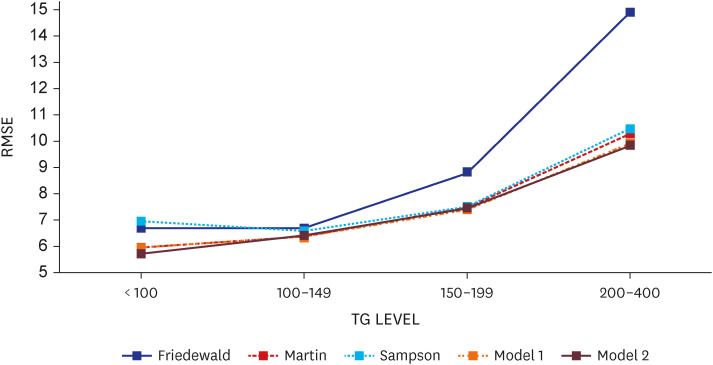
The low-density lipoprotein cholesterol value estimated using the estimation formula and the actual low-density lipoprotein cholesterol value were compared using the root mean squared error. When the triglyceride level was less than 400 mg/dL, the root mean squared of the model 1 was 7.96, the lowest compared to other equations, and the model 2 was 7.82. The degree of misclassification was checked according to the NECP ATP III 6 categories. As a result, the misclassification rate of the model 1 was the lowest at 18.9%, and Weighted Kappa was the highest at 0.919 (0.003), which means it significantly reduced the underestimation rate shown in other existing estimation equations. Root mean square error was also compared according to the change in triglycerides level. As the triglycerides level increased, the root mean square error showed an increasing trend in all equations, but it was confirmed that the model 1 was the lowest compared to other equations.
Conclusion
The newly proposed low-density lipoprotein cholesterol estimation equation showed significantly improved performance compared to the 12 existing estimation equations. The use of representative samples and external verification is required for more sophisticated estimates in the future.
Keyword
Figure
Reference
-
1. Silverman MG, Ference BA, Im K, Wiviott SD, Giugliano RP, Grundy SM, et al. Association between lowering LDL-C and cardiovascular risk reduction among different therapeutic interventions: a systematic review and meta-analysis. JAMA. 2016; 316(12):1289–1297. PMID: 27673306.2. Soran H, Dent R, Durrington P. Evidence-based goals in LDL-C reduction. Clin Res Cardiol. 2017; 106(4):237–248. PMID: 28124099.3. Tremblay AJ, Morrissette H, Gagné JM, Bergeron J, Gagné C, Couture P. Validation of the Friedewald formula for the determination of low-density lipoprotein cholesterol compared with β-quantification in a large population. Clin Biochem. 2004; 37(9):785–790. PMID: 15329317.4. Sampson M, Ling C, Sun Q, Harb R, Ashmaig M, Warnick R, et al. A new equation for calculation of low-density lipoprotein cholesterol in patients with normolipidemia and/or hypertriglyceridemia. JAMA Cardiol. 2020; 5(5):540–548. PMID: 32101259.5. Choi R, Park MJ, Oh Y, Kim SH, Lee SG, Lee EH. Validation of multiple equations for estimating low-density lipoprotein cholesterol levels in Korean adults. Lipids Health Dis. 2021; 20(1):111. PMID: 34544435.6. Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972; 18(6):499–502. PMID: 4337382.7. Grundy SM, Stone NJ, Bailey AL, Beam C, Birtcher KK, Blumenthal RS, et al. 2018 AHA/ACC/AACVPR/AAPA/ABC/ACPM/ADA/AGS/APhA/ASPC/NLA/PCNA guideline on the management of blood cholesterol: executive summary: a report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. J Am Coll Cardiol. 2019; 73(24):3168–3209. PMID: 30423391.8. Martin SS, Blaha MJ, Elshazly MB, Toth PP, Kwiterovich PO, Blumenthal RS, et al. Comparison of a novel method vs the Friedewald equation for estimating low-density lipoprotein cholesterol levels from the standard lipid profile. JAMA. 2013; 310(19):2061–2068. PMID: 24240933.9. Meeusen JW, Lueke AJ, Jaffe AS, Saenger AK. Validation of a proposed novel equation for estimating LDL cholesterol. Clin Chem. 2014; 60(12):1519–1523. PMID: 25336719.10. Song Y, Lee HS, Baik SJ, Jeon S, Han D, Choi SY, et al. Comparison of the effectiveness of Martin’s equation, Friedewald’s equation, and a Novel equation in low-density lipoprotein cholesterol estimation. Sci Rep. 2021; 11(1):13545. PMID: 34188076.11. Hata Y, Nakajima K. Application of Friedewald’s LDL-cholesterol estimation formula to serum lipids in the Japanese population. Jpn Circ J. 1986; 50(12):1191–1200. PMID: 3469420.12. Kang M, Kim J, Lee SY, Kim K, Yoon J, Ki H. Martin’s equation as the most suitable method for estimation of low-density lipoprotein cholesterol levels in Korean adults. Korean J Fam Med. 2017; 38(5):263–269. PMID: 29026486.13. Lee J, Jang S, Son H. Validation of the martin method for estimating low-density lipoprotein cholesterol levels in Korean adults: findings from the Korea National Health and Nutrition Examination Survey, 2009–2011. PLoS One. 2016; 11(1):e0148147. PMID: 26824910.14. Kwon YJ, Lee H, Baik SJ, Chang HJ, Lee JW. Comparison of a machine learning method and various equations for estimating low-density lipoprotein cholesterol in Korean populations. Front Cardiovasc Med. 2022; 9:824574. PMID: 35224055.15. Singh G, Hussain Y, Xu Z, Sholle E, Michalak K, Dolan K, et al. Comparing a novel machine learning method to the Friedewald formula and Martin-Hopkins equation for low-density lipoprotein estimation. PLoS One. 2020; 15(9):e0239934. PMID: 32997716.16. Tsigalou C, Panopoulou M, Papadopoulos C, Karvelas A, Tsairidis D, Anagnostopoulos K. Estimation of low-density lipoprotein cholesterol by machine learning methods. Clin Chim Acta. 2021; 517:108–116. PMID: 33667481.17. Lee T, Kim J, Uh Y, Lee H. Deep neural network for estimating low density lipoprotein cholesterol. Clin Chim Acta. 2019; 489:35–40. PMID: 30448282.18. DeLong DM, DeLong ER, Wood PD, Lippel K, Rifkind BM. A comparison of methods for the estimation of plasma low- and very low-density lipoprotein cholesterol. The Lipid Research Clinics Prevalence Study. JAMA. 1986; 256(17):2372–2377. PMID: 3464768.19. Rao A, Parker AH, el-Sheroni NA, Babelly MM. Calculation of low-density lipoprotein cholesterol with use of triglyceride/cholesterol ratios in lipoproteins compared with other calculation methods. Clin Chem. 1988; 34(12):2532–2534. PMID: 3197296.20. Hattori Y, Suzuki M, Tsushima M, Yoshida M, Tokunaga Y, Wang Y, et al. Development of approximate formula for LDL-chol, LDL-apo B and LDL-chol/LDL-apo B as indices of hyperapobetalipoproteinemia and small dense LDL. Atherosclerosis. 1998; 138(2):289–299. PMID: 9690912.21. Anandaraja S, Narang R, Godeswar R, Laksmy R, Talwar KK. Low-density lipoprotein cholesterol estimation by a new formula in Indian population. Int J Cardiol. 2005; 102(1):117–120. PMID: 15939107.22. Puavilai W, Laorugpongse D, Deerochanawong C, Muthapongthavorn N, Srilert P. The accuracy in using modified Friedewald equation to calculate LDL from non-fast triglyceride: a pilot study. J Med Assoc Thai. 2009; 92(2):182–187. PMID: 19253792.23. Vujovic A, Kotur-Stevuljevic J, Spasic S, Bujisic N, Martinovic J, Vujovic M, et al. Evaluation of different formulas for LDL-C calculation. Lipids Health Dis. 2010; 9(1):27. PMID: 20219094.24. Chen Y, Zhang X, Pan B, Jin X, Yao H, Chen B, et al. A modified formula for calculating low-density lipoprotein cholesterol values. Lipids Health Dis. 2010; 9(1):52. PMID: 20487572.25. de Cordova CM, de Cordova MM. A new accurate, simple formula for LDL-cholesterol estimation based on directly measured blood lipids from a large cohort. Ann Clin Biochem. 2013; 50(Pt 1):13–19. PMID: 23108766.26. Chen T, Guestrin C. Xgboost: a scalable tree boosting system. In : KDD '16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining; August 13-17, 2016; Long Beach, CA, USA. San Francisco, CA, USA: Association for Computing Machinery;2016. p. 785–794.27. Fleiss JL, Levin B, Paik MC. Statistical Methods for Rates and Proportions. 3rd ed. New York, NY, USA: John Wiley & Sons;2003.28. Martin SS, Giugliano RP, Murphy SA, Wasserman SM, Stein EA, Ceška R, et al. Comparison of low-density lipoprotein cholesterol assessment by Martin/Hopkins estimation, Friedewald estimation, and preparative ultracentrifugation: insights from the FOURIER trial. JAMA Cardiol. 2018; 3(8):749–753. PMID: 29898218.29. KDCA. Korea National Health and Nutrition Examination Survey items by year. Updated 2022. Accessed June 7, 2022. https://knhanes.kdca.go.kr/knhanes/sub02/sub02_03.do .30. Kim S, Park E. Differences in height, weight, BMI, and obesity rate between 2018 Community Health and Korea National Health and Nutrition Examination Surveys. J Health Inform Stat. 2020; 45(3):281–287.31. Nordestgaard BG, Langsted A, Mora S, Kolovou G, Baum H, Bruckert E, et al. Fasting is not routinely required for determination of a lipid profile: clinical and laboratory implications including flagging at desirable concentration cut-points-a joint consensus statement from the European Atherosclerosis Society and European Federation of Clinical Chemistry and Laboratory Medicine. Eur Heart J. 2016; 37(25):1944–1958. PMID: 27122601.32. Pallazola VA, Quispe R, Elshazly MB, Vakil R, Sathiyakumar V, Jones SR, et al. Time to make a change: assessing LDL-C accurately in the era of modern pharmacotherapeutics and precision medicine. Curr Cardiovasc Risk Rep. 2018; 12(11):1–7.
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Martin's Equation as the Most Suitable Method for Estimation of Low-Density Lipoprotein Cholesterol Levels in Korean Adults
- Intuitive Modification of the Friedewald Formula for Calculation of LDL-Cholesterol
- Recent Guidelines on the Management of Blood Cholesterol: 2013 ACC/AHA Guidelines and 2014 NICE Draft Guidelines
- Cholesterol Lowering Therapy in Coronary Artery Disease: With Particular Reference to Statins
- Should We Intensify Statin Management in ACS Patients with Very Low LDL Cholesterol Levels?