Korean J Sports Med.
2022 Sep;40(3):151-169. 10.5763/kjsm.2022.40.3.151.
From Barbells to Brawns: The Physiology of Resistance Exercise and Skeletal Muscle Growth
- Affiliations
-
- 1Department of Exercise Science, David B. Falk College of Sport and Human Dynamics, Syracuse University, Syracuse, New York, NY, USA
- 2Department of Biology, College of Arts and Sciences, Syracuse University, Syracuse, New York, NY, USA
- KMID: 2532893
- DOI: http://doi.org/10.5763/kjsm.2022.40.3.151
Abstract
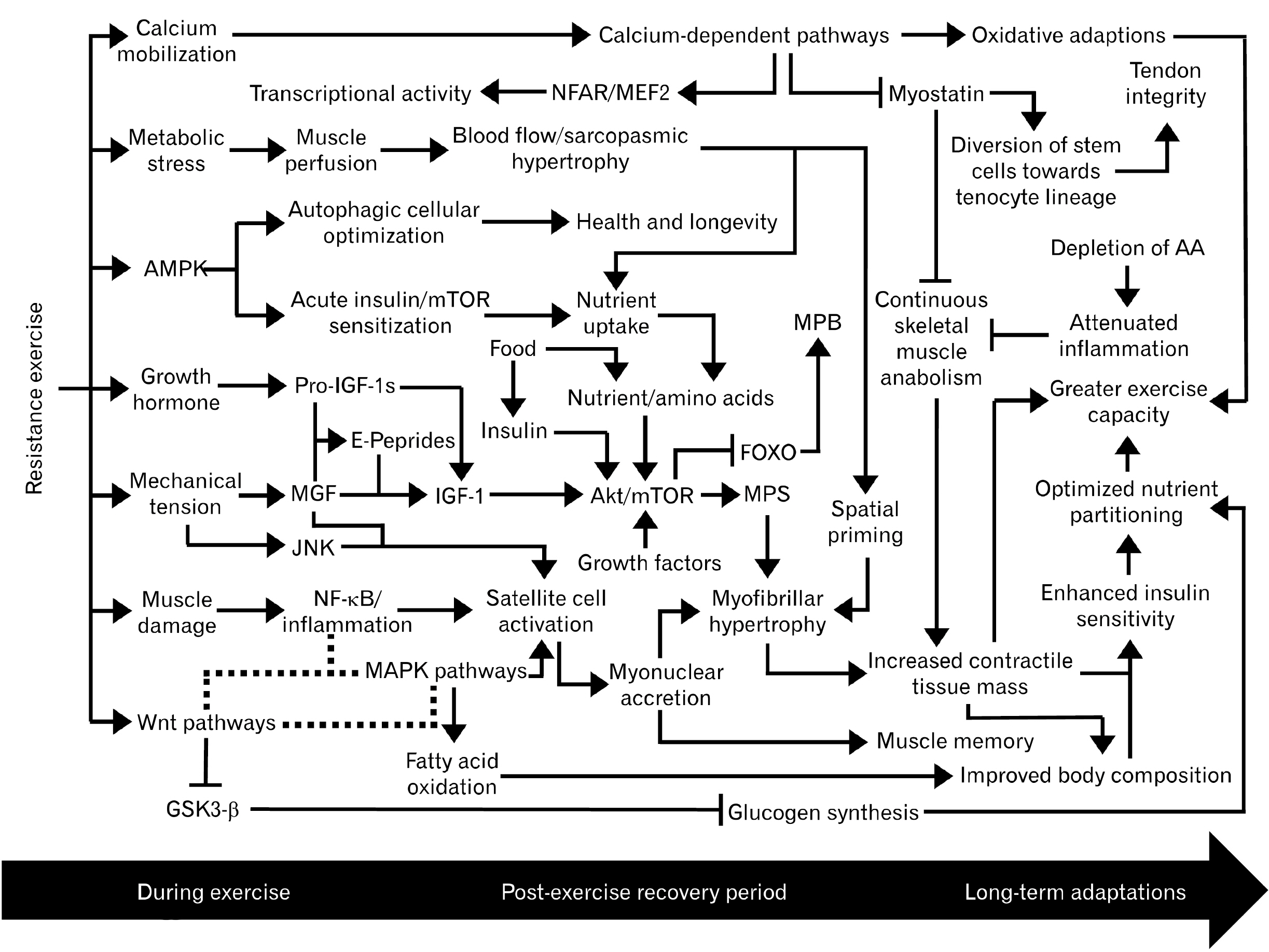
- A complex network of biochemical pathways carries out the process of muscle regeneration/growth following resistance exercise. The initial inflammatory response following muscle damage is primarily mediated by the nuclear factor κ -light-chain-enhancer of activated B cells (NF-κ B), cyclooxygenase enzymes, and prostaglandins. Muscle damage also stimulates the activation, proliferation, differentiation, migration, and fusion of satellite cells onto damaged myofibers, resulting in myofibrillar hypertrophy. The progression of the myogenic lineage is predominantly coordinated by the wingless/integrated family of glycoproteins which engages in crosstalk with NF-κ B and the mitogen-activated protein kinase (MAPK)/extracellular signaling-regulated kinase network. The MAPK cascade is essential for mechanotransduction, the process of converting mechanical stimuli into biochemical responses such as accelerated protein synthesis and satellite cell activation. Muscle protein synthesis is primarily governed by the insulin-like growth factor 1/phosphoinositide 3-kinase/protein kinase B/mammalian target of rapamycin pathway. Several calcium-dependent pathways are also integrated into the process of myogenesis and influence skeletal muscle plasticity. These dynamic interactions are part of the anabolic priming by resistance exercise effect, which defines resistance exercise as an acute catabolic event that potentiates multiple downstream anabolic pathways. Plateaus in muscle growth are attributed to deteriorating inflammatory signaling with repeated bouts of muscle damage as well as increasing thresholds for continuous adaptations, which ultimately become unreachable beyond a certain point. The physiological ceiling of skeletal muscle mass is also credited to myostatin. However, recent discoveries suggest the role of myostatin is not limited to preventing excessive skeletal muscle hypertrophy.
Figure
Reference
-
1. Ryan AS. 2010; Exercise in aging: its important role in mortality, obesity and insulin resistance. Aging Health. 6:551–63. DOI: 10.2217/ahe.10.46. PMID: 21359160. PMCID: PMC3042702.
Article2. Kramer HF, Goodyear LJ. 2007; Exercise, MAPK, and NF-kappaB signaling in skeletal muscle. J Appl Physiol (1985). 103:388–95. DOI: 10.1152/japplphysiol.00085.2007. PMID: 17303713.3. Kraemer WJ, Ratamess NA. 2005; Hormonal responses and adaptations to resistance exercise and training. Sports Med. 35:339–61. DOI: 10.2165/00007256-200535040-00004. PMID: 15831061.
Article4. Schoenfeld BJ. 2010; The mechanisms of muscle hypertrophy and their application to resistance training. J Strength Cond Res. 24:2857–72. DOI: 10.1519/JSC.0b013e3181e840f3. PMID: 20847704.
Article5. Schoenfeld BJ, Contreras B. 2014; The muscle pump: potential mechanisms and applications for enhancing hypertrophic adaptations. Strength Cond J. 36:21–5. DOI: 10.1519/SSC.0000000000000021.6. Yin H, Price F, Rudnicki MA. 2013; Satellite cells and the muscle stem cell niche. Physiol Rev. 93:23–67. DOI: 10.1152/physrev.00043.2011. PMID: 23303905. PMCID: PMC4073943.
Article7. Tesch PA. 1988; Skeletal muscle adaptations consequent to long-term heavy resistance exercise. Med Sci Sports Exerc. 20(5 Suppl):S132–4. DOI: 10.1249/00005768-198810001-00008. PMID: 3057312.
Article8. Schmidt M, Schüler SC, Hüttner SS, von Eyss B, von Maltzahn J. 2019; Adult stem cells at work: regenerating skeletal muscle. Cell Mol Life Sci. 76:2559–70. DOI: 10.1007/s00018-019-03093-6. PMID: 30976839. PMCID: PMC6586695.
Article9. Adams GR. 2006; Satellite cell proliferation and skeletal muscle hypertrophy. Appl Physiol Nutr Metab. 31:782–90. DOI: 10.1139/h06-053. PMID: 17213900.
Article10. Park J, Mcllvain V, Rosenberg J, Donovan L, Desai P, Kim JY. 2022; The mechanisms of anabolic steroids, selective androgen receptor modulators and myostatin inhibitors. Korean J Sports Med. 40:67–85. DOI: 10.5763/kjsm.2022.40.2.67.
Article11. Laplante M, Sabatini DM. 2012; mTOR signaling in growth control and disease. Cell. 149:274–93. DOI: 10.1016/j.cell.2012.03.017. PMID: 22500797. PMCID: PMC3331679.
Article12. Saxton RA, Sabatini DM. 2017; mTOR signaling in growth, metabolism, and disease. Cell. 168:960–76. DOI: 10.1016/j.cell.2017.02.004. PMID: 28283069. PMCID: PMC5394987.
Article13. Seale P, Asakura A, Rudnicki MA. 2001; The potential of muscle stem cells. Dev Cell. 1:333–42. DOI: 10.1016/S1534-5807(01)00049-1.
Article14. Relaix F, Zammit PS. 2012; Satellite cells are essential for skeletal muscle regeneration: the cell on the edge returns centre stage. Development. 139:2845–56. DOI: 10.1242/dev.069088. PMID: 22833472.15. Brook MS, Wilkinson DJ, Smith K, Atherton PJ. 2019; It's not just about protein turnover: the role of ribosomal biogenesis and satellite cells in the regulation of skeletal muscle hypertrophy. Eur J Sport Sci. 19:952–63. DOI: 10.1080/17461391.2019.1569726. PMID: 30741116.
Article16. Murach KA, Englund DA, Dupont-Versteegden EE, McCarthy JJ, Peterson CA. 2018; Myonuclear domain flexibility challenges rigid assumptions on satellite cell contribution to skeletal muscle fiber hypertrophy. Front Physiol. 9:635. DOI: 10.3389/fphys.2018.00635. PMID: 29896117. PMCID: PMC5986879.
Article17. Bruusgaard JC, Johansen IB, Egner IM, Rana ZA, Gundersen K. 2010; Myonuclei acquired by overload exercise precede hypertrophy and are not lost on detraining. Proc Natl Acad Sci U S A. 107:15111–6. DOI: 10.1073/pnas.0913935107. PMID: 20713720. PMCID: PMC2930527.
Article18. Hernández-Hernández JM, García-González EG, Brun CE, Rudnicki MA. 2017; The myogenic regulatory factors, determinants of muscle development, cell identity and regeneration. Semin Cell Dev Biol. 72:10–8. DOI: 10.1016/j.semcdb.2017.11.010.
Article19. Sousa-Victor P, García-Prat L, Muñoz-Cánoves P. 2022; Control of satellite cell function in muscle regeneration and its disruption in ageing. Nat Rev Mol Cell Biol. 23:204–26. DOI: 10.1038/s41580-021-00421-2. PMID: 34663964.20. Haun CT, Vann CG, Roberts BM, Vigotsky AD, Schoenfeld BJ, Roberts MD. 2019; A critical evaluation of the biological construct skeletal muscle hypertrophy: size matters but so does the measurement. Front Physiol. 10:247. DOI: 10.3389/fphys.2019.00247. PMID: 30930796. PMCID: PMC6423469.
Article21. Antonio J, Gonyea WJ. 1993; Skeletal muscle fiber hyperplasia. Med Sci Sports Exerc. 25:1333–45. DOI: 10.1249/00005768-199312000-00004. PMID: 8107539.
Article22. Haun CT, Vann CG, Osburn SC, et al. 2019; Muscle fiber hypertrophy in response to 6 weeks of high-volume resistance training in trained young men is largely attributed to sarcoplasmic hypertrophy. PLoS One. 14:e0215267. DOI: 10.1371/journal.pone.0215267. PMID: 31166954. PMCID: PMC6550381.
Article23. Picard M, Hepple RT, Burelle Y. 2012; Mitochondrial functional specialization in glycolytic and oxidative muscle fibers: tailoring the organelle for optimal function. Am J Physiol Cell Physiol. 302:C629–41. DOI: 10.1152/ajpcell.00368.2011. PMID: 22031602.
Article24. van Wessel T, de Haan A, van der Laarse WJ, Jaspers RT. 2010; The muscle fiber type-fiber size paradox: hypertrophy or oxidative metabolism? Eur J Appl Physiol. 110:665–94. DOI: 10.1007/s00421-010-1545-0. PMID: 20602111. PMCID: PMC2957584.
Article25. de Freitas MC, Gerosa-Neto J, Zanchi NE, Lira FS, Rossi FE. 2017; Role of metabolic stress for enhancing muscle adaptations: practical applications. World J Methodol. 7:46–54. DOI: 10.5662/wjm.v7.i2.46. PMID: 28706859. PMCID: PMC5489423.26. Delp MD, Laughlin MH. 1998; Regulation of skeletal muscle perfusion during exercise. Acta Physiol Scand. 162:411–9. DOI: 10.1046/j.1365-201X.1998.0324e.x. PMID: 9578387.
Article27. Roberts MD, Haun CT, Vann CG, Osburn SC, Young KC. 2020; Sarcoplasmic hypertrophy in skeletal muscle: a scientific "Unicorn" or resistance training adaptation? Front Physiol. 11:816. DOI: 10.3389/fphys.2020.00816. PMID: 32760293. PMCID: PMC7372125.
Article28. Moore DR, Tang JE, Burd NA, Rerecich T, Tarnopolsky MA, Phillips SM. 2009; Differential stimulation of myofibrillar and sarcoplasmic protein synthesis with protein ingestion at rest and after resistance exercise. J Physiol. 587(Pt 4):897–904. DOI: 10.1113/jphysiol.2008.164087. PMID: 19124543. PMCID: PMC2669978.29. Vann CG, Roberson PA, Osburn SC, et al. 2020; Skeletal muscle myofibrillar protein abundance is higher in resistance-trained men, and aging in the absence of training may have an opposite effect. Sports (Basel). 8:7. DOI: 10.3390/sports8010007. PMID: 31936810. PMCID: PMC7022975.
Article30. Douglas J, Pearson S, Ross A, McGuigan M. 2017; Eccentric exercise: physiological characteristics and acute responses. Sports Med. 47:663–75. DOI: 10.1007/s40279-016-0624-8. PMID: 27638040.
Article31. De Souza EO, Lowery RP, Wilson JM, et al. 2016; Effects of arachidonic acid supplementation on acute anabolic signaling and chronic functional performance and body composition adaptations. PLoS One. 11:e0155153. DOI: 10.1371/journal.pone.0155153. PMID: 27182886. PMCID: PMC4868363.
Article32. Schoenfeld BJ. 2012; The use of nonsteroidal anti-inflammatory drugs for exercise-induced muscle damage: implications for skeletal muscle development. Sports Med. 42:1017–28. DOI: 10.1007/BF03262309. PMID: 23013520.
Article33. Costamagna D, Costelli P, Sampaolesi M, Penna F. 2015; Role of inflammation in muscle homeostasis and myogenesis. Mediators Inflamm. 2015:805172. DOI: 10.1155/2015/805172. PMID: 26508819. PMCID: PMC4609834.34. Arnold L, Henry A, Poron F, et al. 2007; Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 204:1057–69. DOI: 10.1084/jem.20070075. PMID: 17485518. PMCID: PMC2118577.
Article35. Markworth JF, Cameron-Smith D. 2013; Arachidonic acid supplementation enhances in vitro skeletal muscle cell growth via a COX-2-dependent pathway. Am J Physiol Cell Physiol. 304:C56–67. DOI: 10.1152/ajpcell.00038.2012. PMID: 23076795.
Article36. Veliça P, Bunce CM. 2008; Prostaglandins in muscle regeneration. J Muscle Res Cell Motil. 29:163–7. DOI: 10.1007/s10974-008-9154-9.
Article37. Korotkova M, Lundberg IE. 2014; The skeletal muscle arachidonic acid cascade in health and inflammatory disease. Nat Rev Rheumatol. 10:295–303. DOI: 10.1038/nrrheum.2014.2. PMID: 24468934.38. Trappe TA, Liu SZ. 2013; Effects of prostaglandins and COX-inhibiting drugs on skeletal muscle adaptations to exercise. J Appl Physiol (1985). 115:909–19. DOI: 10.1152/japplphysiol.00061.2013. PMID: 23539318. PMCID: PMC3764617.
Article39. Palmer RM. 1990; Prostaglandins and the control of muscle protein synthesis and degradation. Prostaglandins Leukot Essent Fatty Acids. 39:95–104. DOI: 10.1016/0952-3278(90)90017-F.
Article40. Sanchez AM, Bernardi H, Py G, Candau RB. 2014; Autophagy is essential to support skeletal muscle plasticity in response to endurance exercise. Am J Physiol Regul Integr Comp Physiol. 307:R956–69. DOI: 10.1152/ajpregu.00187.2014. PMID: 25121614.
Article41. Otis JS, Burkholder TJ, Pavlath GK. 2005; Stretch-induced myoblast proliferation is dependent on the COX2 pathway. Exp Cell Res. 310:417–25. DOI: 10.1016/j.yexcr.2005.08.009. PMID: 16168411.
Article42. Hoxha M. 2019; Duchenne muscular dystrophy: focus on arachidonic acid metabolites. Biomed Pharmacother. 110:796–802. DOI: 10.1016/j.biopha.2018.12.034. PMID: 30554118.43. Webster JM, Kempen LJ, Hardy RS, Langen RC. 2020; Inflammation and skeletal muscle wasting during cachexia. Front Physiol. 11:597675. DOI: 10.3389/fphys.2020.597675. PMID: 33329046. PMCID: PMC7710765.44. Woods JA, Wilund KR, Martin SA, Kistler BM. 2012; Exercise, inflammation and aging. Aging Dis. 3:130–40.45. Cheung K, Hume P, Maxwell L. 2003; Delayed onset muscle soreness: treatment strategies and performance factors. Sports Med. 33:145–64. DOI: 10.2165/00007256-200333020-00005. PMID: 12617692.46. Straughn AR, Hindi SM, Xiong G, Kumar A. 2019; Canonical NF-κB signaling regulates satellite stem cell homeostasis and function during regenerative myogenesis. J Mol Cell Biol. 11:53–66. DOI: 10.1093/jmcb/mjy053. PMID: 30239789.
Article47. Vella L, Markworth JF, Peake JM, Snow RJ, Cameron-Smith D, Russell AP. 2014; Ibuprofen supplementation and its effects on NF-κB activation in skeletal muscle following resistance exercise. Physiol Rep. 2:e12172. DOI: 10.14814/phy2.12172. PMID: 25344476. PMCID: PMC4254097.48. Li H, Malhotra S, Kumar A. 2008; Nuclear factor-kappa B signaling in skeletal muscle atrophy. J Mol Med (Berl). 86:1113–26. DOI: 10.1007/s00109-008-0373-8. PMID: 18574572. PMCID: PMC2597184.
Article49. Ma B, Hottiger MO. 2016; Crosstalk between Wnt/β-catenin and NF-κB signaling pathway during inflammation. Front Immunol. 7:378. DOI: 10.3389/fimmu.2016.00378. PMID: 27713747. PMCID: PMC5031610.
Article50. Komiya Y, Habas R. 2008; Wnt signal transduction pathways. Organogenesis. 4:68–75. DOI: 10.4161/org.4.2.5851. PMID: 19279717. PMCID: PMC2634250.
Article51. von Maltzahn J, Bentzinger CF, Rudnicki MA. 2011; Wnt7a-Fzd7 signalling directly activates the Akt/mTOR anabolic growth pathway in skeletal muscle. Nat Cell Biol. 14:186–91. DOI: 10.1038/ncb2404. PMID: 22179044. PMCID: PMC3271181.
Article52. Castilho RM, Squarize CH, Chodosh LA, Williams BO, Gutkind JS. 2009; mTOR mediates Wnt-induced epidermal stem cell exhaustion and aging. Cell Stem Cell. 5:279–89. DOI: 10.1016/j.stem.2009.06.017. PMID: 19733540. PMCID: PMC2939833.
Article53. Sethi JK, Vidal-Puig A. 2010; Wnt signalling and the control of cellular metabolism. Biochem J. 427:1–17. DOI: 10.1042/BJ20091866. PMID: 20226003. PMCID: PMC4301310.
Article54. von Maltzahn J, Chang NC, Bentzinger CF, Rudnicki MA. 2012; Wnt signaling in myogenesis. Trends Cell Biol. 22:602–9. DOI: 10.1016/j.tcb.2012.07.008. PMID: 22944199. PMCID: PMC3479319.
Article55. Bornens M. 2008; Organelle positioning and cell polarity. Nat Rev Mol Cell Biol. 9:874–86. DOI: 10.1038/nrm2524. PMID: 18946476.
Article56. Le Grand F, Jones AE, Seale V, Scimè A, Rudnicki MA. 2009; Wnt7a activates the planar cell polarity pathway to drive the symmetric expansion of satellite stem cells. Cell Stem Cell. 4:535–47. DOI: 10.1016/j.stem.2009.03.013. PMID: 19497282. PMCID: PMC2743383.
Article57. Huraskin D, Eiber N, Reichel M, et al. 2016; Wnt/β-catenin signaling via Axin2 is required for myogenesis and, together with YAP/Taz and Tead1, active in IIa/IIx muscle fibers. Development. 143:3128–42. DOI: 10.1242/dev.139907. PMID: 27578179.
Article58. Ramakrishnan AB, Cadigan KM. 2017; Wnt target genes and where to find them. F1000Res. 6:746. DOI: 10.12688/f1000research.11034.1. PMID: 28649368. PMCID: PMC5464219.
Article59. Tidball JG, Welc SS. 2015; Macrophage-derived IGF-1 is a potent coordinator of myogenesis and inflammation in regenerating muscle. Mol Ther. 23:1134–5. DOI: 10.1038/mt.2015.97. PMID: 26122828. PMCID: PMC4817792.
Article60. Philippou A, Maridaki M, Pneumaticos S, Koutsilieris M. 2014; The complexity of the IGF1 gene splicing, posttranslational modification and bioactivity. Mol Med. 20:202–14. DOI: 10.2119/molmed.2014.00011. PMID: 24637928. PMCID: PMC4022784.
Article61. Osher E, Macaulay VM. 2019; Therapeutic targeting of the IGF axis. Cells. 8:895. DOI: 10.3390/cells8080895. PMID: 31416218. PMCID: PMC6721736.
Article62. Pasiakos SM. 2012; Exercise and amino acid anabolic cell signaling and the regulation of skeletal muscle mass. Nutrients. 4:740–58. DOI: 10.3390/nu4070740. PMID: 22852061. PMCID: PMC3407992.
Article63. Brisson BK, Barton ER. 2013; New modulators for IGF-I activity within IGF-I processing products. Front Endocrinol (Lausanne). 4:42. DOI: 10.3389/fendo.2013.00042. PMID: 23543904. PMCID: PMC3608916.
Article64. Ascenzi F, Barberi L, Dobrowolny G, et al. 2019; Effects of IGF-1 isoforms on muscle growth and sarcopenia. Aging Cell. 18:e12954. DOI: 10.1111/acel.12954. PMID: 30953403. PMCID: PMC6516183.
Article65. Philippou A, Papageorgiou E, Bogdanis G, et al. 2009; Expression of IGF-1 isoforms after exercise-induced muscle damage in humans: characterization of the MGF E peptide actions in vitro. In Vivo. 23:567–75.66. Matheny RW Jr, Nindl BC, Adamo ML. 2010; Minireview: Mechano-growth factor: a putative product of IGF-I gene expression involved in tissue repair and regeneration. Endocrinology. 151:865–75. DOI: 10.1210/en.2009-1217. PMID: 20130113. PMCID: PMC2840678.
Article67. Aboalola D, Han VK. 2017; Different effects of insulin-like growth factor-1 and insulin-like growth factor-2 on myogenic differentiation of human mesenchymal stem cells. Stem Cells Int. 2017:8286248. DOI: 10.1155/2017/8286248. PMID: 29387091. PMCID: PMC5745708.
Article68. Dreyer HC, Fujita S, Cadenas JG, Chinkes DL, Volpi E, Rasmussen BB. 2006; Resistance exercise increases AMPK activity and reduces 4E-BP1 phosphorylation and protein synthesis in human skeletal muscle. J Physiol. 576(Pt 2):613–24. DOI: 10.1113/jphysiol.2006.113175. PMID: 16873412. PMCID: PMC1890364.
Article69. Frost RA, Lang CH. 2007; Protein kinase B/Akt: a nexus of growth factor and cytokine signaling in determining muscle mass. J Appl Physiol (1985). 103:378–87. DOI: 10.1152/japplphysiol.00089.2007. PMID: 17332274.
Article70. Laplante M, Sabatini DM. 2009; mTOR signaling at a glance. J Cell Sci. 122(Pt 20):3589–94. DOI: 10.1242/jcs.051011. PMID: 19812304. PMCID: PMC2758797.
Article71. Tremblay F, Marette A. 2001; Amino acid and insulin signaling via the mTOR/p70 S6 kinase pathway: a negative feedback mechanism leading to insulin resistance in skeletal muscle cells. J Biol Chem. 276:38052–60. DOI: 10.1074/jbc.M106703200. PMID: 11498541.72. White JP, Gao S, Puppa MJ, Sato S, Welle SL, Carson JA. 2013; Testosterone regulation of Akt/mTORC1/FoxO3a signaling in skeletal muscle. Mol Cell Endocrinol. 365:174–86. DOI: 10.1016/j.mce.2012.10.019. PMID: 23116773. PMCID: PMC3529800.
Article73. Kjøbsted R, Hingst JR, Fentz J, et al. 2018; AMPK in skeletal muscle function and metabolism. FASEB J. 32:1741–77. DOI: 10.1096/fj.201700442R. PMID: 29242278. PMCID: PMC5945561.
Article74. Zhang P, Liang X, Shan T, et al. 2015; mTOR is necessary for proper satellite cell activity and skeletal muscle regeneration. Biochem Biophys Res Commun. 463:102–8. DOI: 10.1016/j.bbrc.2015.05.032. PMID: 25998386. PMCID: PMC4484853.75. Dibble CC, Cantley LC. 2015; Regulation of mTORC1 by PI3K signaling. Trends Cell Biol. 25:545–55. DOI: 10.1016/j.tcb.2015.06.002. PMID: 26159692. PMCID: PMC4734635.
Article76. Bohé J, Low JF, Wolfe RR, Rennie MJ. 2001; Latency and duration of stimulation of human muscle protein synthesis during continuous infusion of amino acids. J Physiol. 532(Pt 2):575–9. DOI: 10.1111/j.1469-7793.2001.0575f.x. PMID: 11306673. PMCID: PMC2278544.77. Atherton PJ, Etheridge T, Watt PW, et al. 2010; Muscle full effect after oral protein: time-dependent concordance and discordance between human muscle protein synthesis and mTORC1 signaling. Am J Clin Nutr. 92:1080–8. DOI: 10.3945/ajcn.2010.29819. PMID: 20844073.
Article78. Norton LE, Wilson GJ. 2009; Optimal protein intake to maximize muscle protein synthesis: examinations of optimal meal protein intake and frequency for athletes. Agro Food Ind Hi Tech. 20:54–7.79. Areta JL, Burke LM, Ross ML, et al. 2013; Timing and distribution of protein ingestion during prolonged recovery from resistance exercise alters myofibrillar protein synthesis. J Physiol. 591:2319–31. DOI: 10.1113/jphysiol.2012.244897. PMID: 23459753. PMCID: PMC3650697.
Article80. Slater GJ, Dieter BP, Marsh DJ, Helms ER, Shaw G, Iraki J. 2019; Is an energy surplus required to maximize skeletal muscle hypertrophy associated with resistance training. Front Nutr. 6:131. DOI: 10.3389/fnut.2019.00131. PMID: 31482093. PMCID: PMC6710320.
Article81. Zanchi NE, Gerlinger-Romero F, Guimarães-Ferreira L, et al. 2011; HMB supplementation: clinical and athletic performance-related effects and mechanisms of action. Amino Acids. 40:1015–25. DOI: 10.1007/s00726-010-0678-0. PMID: 20607321.
Article82. Hector AJ, Phillips SM. 2018; Protein recommendations for weight loss in elite athletes: a focus on body composition and performance. Int J Sport Nutr Exerc Metab. 28:170–7. DOI: 10.1123/ijsnem.2017-0273. PMID: 29182451.83. Gallagher PM, Touchberry CD, Teson K, McCabe E, Tehel M, Wacker MJ. 2013; Effects of an acute bout of resistance exercise on fiber-type specific to GLUT4 and IGF-1R expression. Appl Physiol Nutr Metab. 38:581–6. DOI: 10.1139/apnm-2012-0301. PMID: 23668768.84. Zhang W, Liu HT. 2002; MAPK signal pathways in the regulation of cell proliferation in mammalian cells. Cell Res. 12:9–18. DOI: 10.1038/sj.cr.7290105. PMID: 11942415.85. Mendoza MC, Er EE, Blenis J. 2011; The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 36:320–8. DOI: 10.1016/j.tibs.2011.03.006. PMID: 21531565. PMCID: PMC3112285.
Article86. Al-Shanti N, Stewart CE. 2009; Ca2+/calmodulin-dependent transcriptional pathways: potential mediators of skeletal muscle growth and development. Biol Rev Camb Philos Soc. 84:637–52. DOI: 10.1111/j.1469-185X.2009.00090.x. PMID: 19725819.87. Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. 2000; Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J Biol Chem. 275:4545–8. DOI: 10.1074/jbc.275.7.4545. PMID: 10671477.
Article88. Methenitis S. 2018; A brief review on concurrent training: from laboratory to the field. Sports (Basel). 6:127. DOI: 10.3390/sports6040127. PMID: 30355976. PMCID: PMC6315763.
Article89. Lee SJ, Lee YS, Zimmers TA, et al. 2010; Regulation of muscle mass by follistatin and activins. Mol Endocrinol. 24:1998–2008. DOI: 10.1210/me.2010-0127. PMID: 20810712. PMCID: PMC2954636.
Article90. Lee SJ, Huynh TV, Lee YS, et al. 2012; Role of satellite cells versus myofibers in muscle hypertrophy induced by inhibition of the myostatin/activin signaling pathway. Proc Natl Acad Sci U S A. 109:E2353–60. DOI: 10.1073/pnas.1206410109. PMID: 22869749. PMCID: PMC3435227.
Article91. Rodriguez J, Vernus B, Chelh I, et al. 2014; Myostatin and the skeletal muscle atrophy and hypertrophy signaling pathways. Cell Mol Life Sci. 71:4361–71. DOI: 10.1007/s00018-014-1689-x. PMID: 25080109.
Article92. Lee YS, Lee SJ. 2013; Regulation of GDF-11 and myostatin activity by GASP-1 and GASP-2. Proc Natl Acad Sci U S A. 110:E3713–22. DOI: 10.1073/pnas.1309907110. PMID: 24019467. PMCID: PMC3785741.
Article93. Zhu X, Topouzis S, Liang LF, Stotish RL. 2004; Myostatin signaling through Smad2, Smad3 and Smad4 is regulated by the inhibitory Smad7 by a negative feedback mechanism. Cytokine. 26:262–72. DOI: 10.1016/j.cyto.2004.03.007. PMID: 15183844.
Article94. Walker RG, Poggioli T, Katsimpardi L, et al. 2016; Biochemistry and biology of GDF11 and myostatin: similarities, differences, and questions for future investigation. Circ Res. 118:1125–42. DOI: 10.1161/CIRCRESAHA.116.308391. PMID: 27034275. PMCID: PMC4818972.95. Mendias CL, Lynch EB, Gumucio JP, et al. 2015; Changes in skeletal muscle and tendon structure and function following genetic inactivation of myostatin in rats. J Physiol. 593:2037–52. DOI: 10.1113/jphysiol.2014.287144. PMID: 25640143. PMCID: PMC4405758.
Article96. Guo T, Jou W, Chanturiya T, Portas J, Gavrilova O, McPherron AC. 2009; Myostatin inhibition in muscle, but not adipose tissue, decreases fat mass and improves insulin sensitivity. PLoS One. 4:e4937. DOI: 10.1371/journal.pone.0004937. PMID: 19295913. PMCID: PMC2654157.
Article97. Schuelke M, Wagner KR, Stolz LE, et al. 2004; Myostatin mutation associated with gross muscle hypertrophy in a child. N Engl J Med. 350:2682–8. DOI: 10.1056/NEJMoa040933. PMID: 15215484.
Article98. Suh J, Lee YS. 2020; Myostatin inhibitors: panacea or predicament for musculoskeletal disorders? J Bone Metab. 27:151–65. DOI: 10.11005/jbm.2020.27.3.151. PMID: 32911580. PMCID: PMC7571243.
Article99. Le W, Yao J. 2017; The effect of myostatin (GDF-8) on proliferation and tenocyte differentiation of rat bone marrow-derived mesenchymal stem cells. J Hand Surg Asian Pac Vol. 22:200–7. DOI: 10.1142/S0218810417500253. PMID: 28506172.100. Welle S, Cardillo A, Zanche M, Tawil R. 2009; Skeletal muscle gene expression after myostatin knockout in mature mice. Physiol Genomics. 38:342–50. DOI: 10.1152/physiolgenomics.00054.2009. PMID: 19509079. PMCID: PMC3774565.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Regulation of exercise-stimulated glucose uptake in skeletal muscle
- Skeletal Muscle Mitochondria and Insulin Resistance: The Role of Exercise
- Exercise/Resistance Training and Muscle Stem Cells
- Effect of Resistance Exercise Training on Mustn1 mRNA Expression in Rat Skeletal Muscle
- Myostatin as a Potential Therapeutic Target for Obesity and Insulin Resistance