J Korean Med Sci.
2021 Dec;36(50):e342. 10.3346/jkms.2021.36.e342.
The Effect of Formula-based Nutritional Treatment on Colitis in a Murine Model
- Affiliations
-
- 1Division of Gastroenterology, Hepatology and Nutrition, Department of Pediatrics, Yonsei University College of Medicine, Severance Fecal Microbiota Transplantation Center, Severance Hospital, Seoul, Korea
- 2Department of Medicine, Yonsei University College of Medicine, Seoul, Korea
- 3Department of Microbiology and Immunology, Brain Korea 21 Project for Medical Sciences, Yonsei University College of Medicine, Seoul, Korea
- 4Institute for Immunology and Immunological Diseases, Yonsei University College of Medicine, Seoul, Korea
- KMID: 2523562
- DOI: http://doi.org/10.3346/jkms.2021.36.e342
Abstract
- Background
Exclusive enteral nutrition (EEN) induces remission in pediatric Crohn's disease (CD). The exact mechanism of EEN therapy in CD is unknown, but alteration of the intestinal microflora after EEN is thought to affect mucosal healing. To determine the link between EEN therapy and therapeutic efficacy in CD, we established a murine model of dextran sulfate sodium (DSS)-induced colitis and applied EEN therapy.
Methods
Eight-week-old C57BL/6 mice were administered DSS for 4 days to induce colitis, and either normal chow (NC) or EEN was administered for the following 4 days. The mice were grouped according to the feeding pattern after DSS administration: DSS/NC and DSS/ EEN groups. The clinical course was confirmed via daily observation of the weight and stool. Fecal samples were collected and 16sRNA sequencing was used. The mice were sacrificed to confirm colonic histopathology.
Results
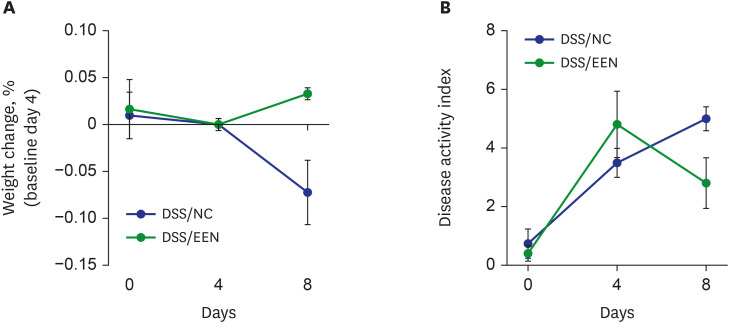
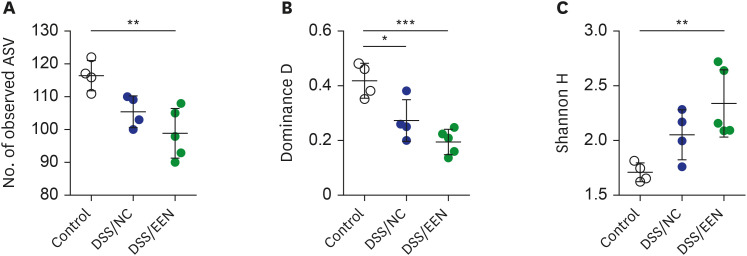
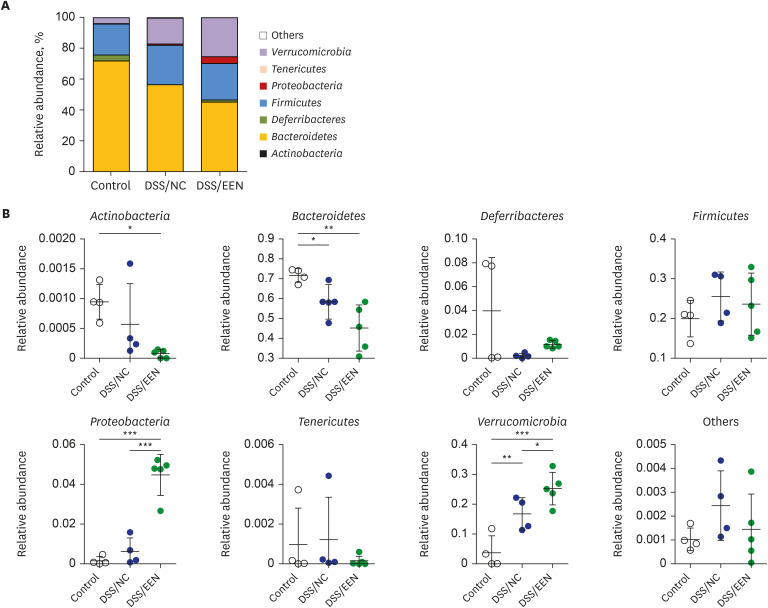
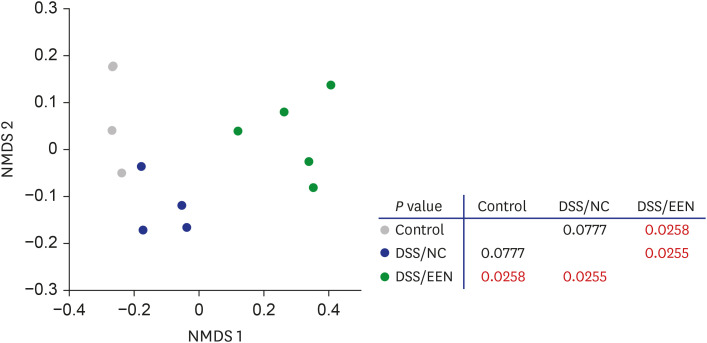
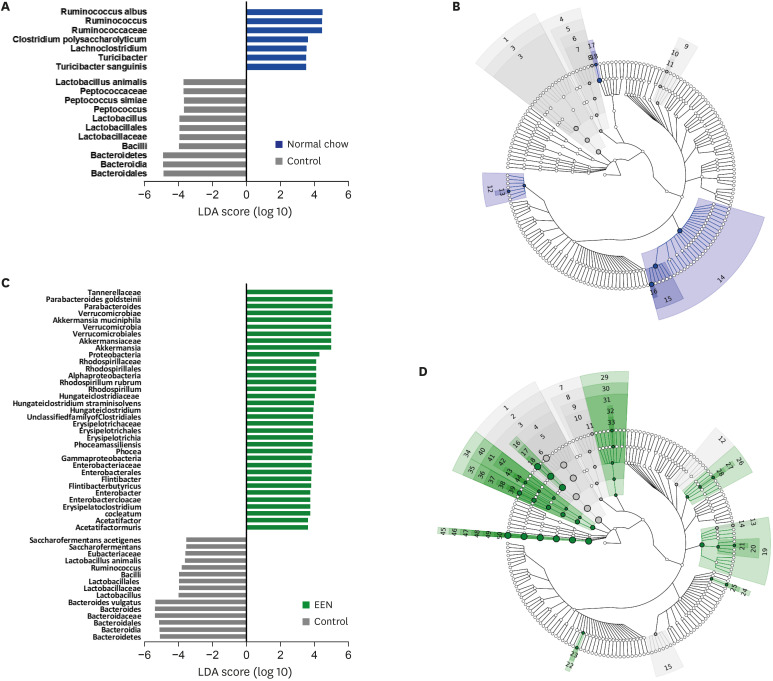
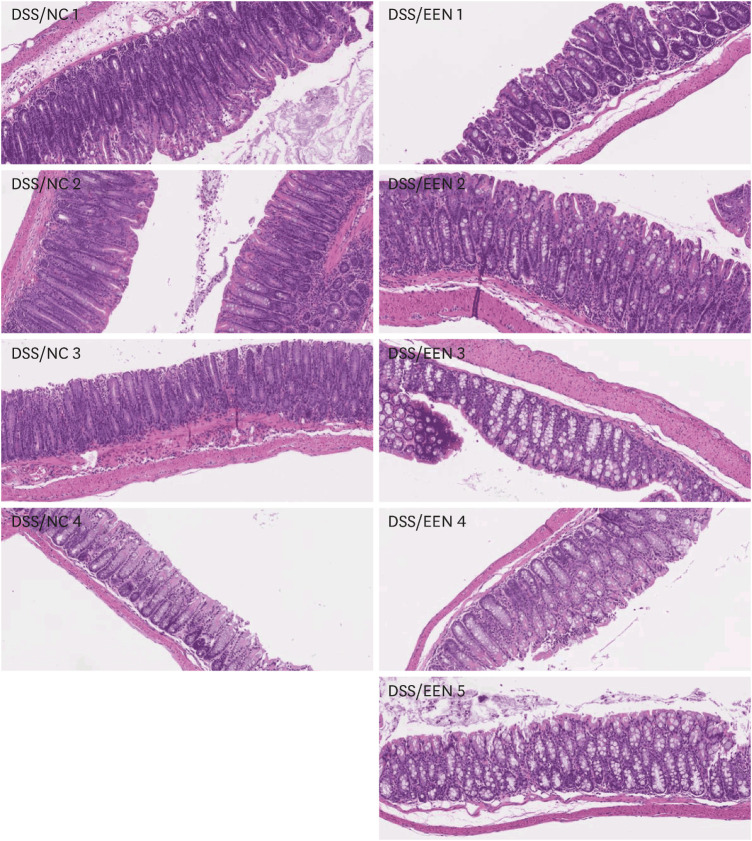
Weight reduction and increase in disease activity were observed as the day progressed for 4 days after DSS administration. There was significant weight recovery and improvement in disease activity in the EEN group compared to that in the NC group. Verrucomicrobia and Proteobacteria abundances tended to increase and Bacteroidetes abundance decreased in the EEN group. In the EEN group, significant changes in the β-diversity of the microbiota were observed. In the analysis of microbiome species, abundances of Akkermansia muciniphila, Clostridium cocleatum, mucin-degrading bacteria, Flintibacter butyricus, and Parabacteroides goldsteinii, which are beneficial microbiota, were significantly increased in the EEN group compared to those in the NC group. More abundant mucins were confirmed in the colonic histopathology of the EEN group. These microbial and histopathological differences suggested that EEN might improve colitis symptoms in a murine colitis model by promoting mucin recycling and subsequently inducing the healing effect of the gut barrier.
Conclusion
EEN showed clinical efficacy in a murine model of colitis. Based on the increase in mucin-degrading bacteria and the pathological increase in mucin production after EEN administration, it can be observed that mucin plays an important role in the therapeutic effect of EEN.
Keyword
Figure
Reference
-
1. Boyapati R, Satsangi J, Ho GT. Pathogenesis of Crohn’s disease. F1000Prime Rep. 2015; 7:44. PMID: 26097717.
Article2. Day AS, Lopez RN. Exclusive enteral nutrition in children with Crohn’s disease. World J Gastroenterol. 2015; 21(22):6809–6816. PMID: 26078556.
Article3. Ruemmele FM, Veres G, Kolho KL, Griffiths A, Levine A, Escher JC, et al. Consensus guidelines of ECCO/ESPGHAN on the medical management of pediatric Crohn’s disease. J Crohns Colitis. 2014; 8(10):1179–1207. PMID: 24909831.
Article4. Adamji M, Day AS. An overview of the role of exclusive enteral nutrition for complicated Crohn’s disease. Intest Res. 2019; 17(2):171–176. PMID: 30508476.
Article5. Bannerjee K, Camacho-Hübner C, Babinska K, Dryhurst KM, Edwards R, Savage MO, et al. Anti-inflammatory and growth-stimulating effects precede nutritional restitution during enteral feeding in Crohn disease. J Pediatr Gastroenterol Nutr. 2004; 38(3):270–275. PMID: 15076624.
Article6. Teahon K, Pearson M, Smith T, Bjarnason I. Alterations in nutritional status and disease activity during treatment of Crohn’s disease with elemental diet. Scand J Gastroenterol. 1995; 30(1):54–60. PMID: 7701251.
Article7. Nahidi L, Leach ST, Mitchell HM, Kaakoush NO, Lemberg DA, Munday JS, et al. Inflammatory bowel disease therapies and gut function in a colitis mouse model. BioMed Res Int. 2013; 2013:909613. PMID: 24027765.
Article8. Nagy-Szakal D, Mir SA, Ross MC, Tatevian N, Petrosino JF, Kellermayer R. Monotonous diets protect against acute colitis in mice: epidemiologic and therapeutic implications. J Pediatr Gastroenterol Nutr. 2013; 56(5):544–550. PMID: 23085891.9. Gatti S, Galeazzi T, Franceschini E, Annibali R, Albano V, Verma AK, et al. Effects of the exclusive enteral nutrition on the microbiota profile of patients with Crohn’s disease: a systematic review. Nutrients. 2017; 9(8):E832. PMID: 28777338.
Article10. Okayasu I, Hatakeyama S, Yamada M, Ohkusa T, Inagaki Y, Nakaya R. A novel method in the induction of reliable experimental acute and chronic ulcerative colitis in mice. Gastroenterology. 1990; 98(3):694–702. PMID: 1688816.
Article11. Perret V, Lev R, Pigman W. Simple method for the preparation of single cell suspensions from normal and tumorous rat colonic mucosa. Gut. 1977; 18(5):382–385. PMID: 873323.
Article12. Murthy SN, Cooper HS, Shim H, Shah RS, Ibrahim SA, Sedergran DJ. Treatment of dextran sulfate sodium-induced murine colitis by intracolonic cyclosporin. Dig Dis Sci. 1993; 38(9):1722–1734. PMID: 8359087.
Article13. Koelink PJ, Wildenberg ME, Stitt LW, Feagan BG, Koldijk M, van ’t Wout AB, et al. Development of reliable, valid and responsive scoring systems for endoscopy and histology in animal models for inflammatory bowel disease. J Crohns Colitis. 2018; 12(7):794–803. PMID: 29608662.
Article14. Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012; 6(8):1621–1624. PMID: 22402401.
Article15. Fadrosh DW, Ma B, Gajer P, Sengamalay N, Ott S, Brotman RM, et al. An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome. 2014; 2(1):6. PMID: 24558975.
Article16. Masella AP, Bartram AK, Truszkowski JM, Brown DG, Neufeld JD. PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics. 2012; 13(1):31. PMID: 22333067.
Article17. Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010; 7(5):335–336. PMID: 20383131.
Article18. Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS, et al. Metagenomic biomarker discovery and explanation. Genome Biol. 2011; 12(6):R60. PMID: 21702898.
Article19. MacLellan A, Moore-Connors J, Grant S, Cahill L, Langille MG, Van Limbergen J. The impact of exclusive enteral nutrition (EEN) on the gut microbiome in Crohn’s disease: a review. Nutrients. 2017; 9(5):447.
Article20. Lionetti P, Callegari ML, Ferrari S, Cavicchi MC, Pozzi E, de Martino M, et al. Enteral nutrition and microflora in pediatric Crohn's disease. JPEN J Parenter Enteral Nutr. 2005; 29(4 Suppl):S173–S175. PMID: 15980280.
Article21. Gerasimidis K, Bertz M, Hanske L, Junick J, Biskou O, Aguilera M, et al. Decline in presumptively protective gut bacterial species and metabolites are paradoxically associated with disease improvement in pediatric Crohn’s disease during enteral nutrition. Inflamm Bowel Dis. 2014; 20(5):861–871. PMID: 24651582.
Article22. Quince C, Ijaz UZ, Loman N, Eren AM, Saulnier D, Russell J, et al. Extensive modulation of the fecal metagenome in children with Crohn’s disease during exclusive enteral nutrition. Am J Gastroenterol. 2015; 110(12):1718–1729. PMID: 26526081.
Article23. D’Argenio V, Precone V, Casaburi G, Miele E, Martinelli M, Staiano A, et al. An altered gut microbiome profile in a child affected by Crohn’s disease normalized after nutritional therapy. Am J Gastroenterol. 2013; 108(5):851–852. PMID: 23644964.24. Diederen K, Li JV, Donachie GE, de Meij TG, de Waart DR, Hakvoort TB, et al. Exclusive enteral nutrition mediates gut microbial and metabolic changes that are associated with remission in children with Crohn’s disease. Sci Rep. 2020; 10(1):18879. PMID: 33144591.
Article25. Derrien M, Vaughan EE, Plugge CM, de Vos WM. Akkermansia muciniphila gen. nov., sp. nov., a human intestinal mucin-degrading bacterium. Int J Syst Evol Microbiol. 2004; 54(Pt 5):1469–1476. PMID: 15388697.26. Boureau H, Decré D, Carlier JP, Guichet C, Bourlioux P. Identification of a Clostridium cocleatum strain involved in an anti-Clostridium difficile barrier effect and determination of its mucin-degrading enzymes. Res Microbiol. 1993; 144(5):405–410. PMID: 7504316.27. Wu TR, Lin CS, Chang CJ, Lin TL, Martel J, Ko YF, et al. Gut commensal Parabacteroides goldsteinii plays a predominant role in the anti-obesity effects of polysaccharides isolated from Hirsutella sinensis . Gut. 2019; 68(2):248–262. PMID: 30007918.28. Gaudier E, Jarry A, Blottière HM, de Coppet P, Buisine MP, Aubert JP, et al. Butyrate specifically modulates MUC gene expression in intestinal epithelial goblet cells deprived of glucose. Am J Physiol Gastrointest Liver Physiol. 2004; 287(6):G1168–G1174. PMID: 15308471.29. Pelaseyed T, Bergström JH, Gustafsson JK, Ermund A, Birchenough GM, Schütte A, et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immunol Rev. 2014; 260(1):8–20. PMID: 24942678.
Article30. Lagkouvardos I, Pukall R, Abt B, Foesel BU, Meier-Kolthoff JP, Kumar N, et al. The Mouse Intestinal Bacterial Collection (miBC) provides host-specific insight into cultured diversity and functional potential of the gut microbiota. Nat Microbiol. 2016; 1(10):16131. PMID: 27670113.
Article31. Dunn KA, Moore-Connors J, MacIntyre B, Stadnyk AW, Thomas NA, Noble A, et al. Early changes in microbial community structure are associated with sustained remission after nutritional treatment of pediatric Crohn’s disease. Inflamm Bowel Dis. 2016; 22(12):2853–2862. PMID: 27805918.
Article32. Everard A, Belzer C, Geurts L, Ouwerkerk JP, Druart C, Bindels LB, et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc Natl Acad Sci U S A. 2013; 110(22):9066–9071. PMID: 23671105.33. Png CW, Lindén SK, Gilshenan KS, Zoetendal EG, McSweeney CS, Sly LI, et al. Mucolytic bacteria with increased prevalence in IBD mucosa augment in vitro utilization of mucin by other bacteria. Am J Gastroenterol. 2010; 105(11):2420–2428. PMID: 20648002.
Article34. Swidsinski A, Dörffel Y, Loening-Baucke V, Theissig F, Rückert JC, Ismail M, et al. Acute appendicitis is characterised by local invasion with Fusobacterium nucleatum/necrophorum . Gut. 2011; 60(1):34–40. PMID: 19926616.35. Ó Cuív P, de Wouters T, Giri R, Mondot S, Smith WJ, Blottière HM, et al. The gut bacterium and pathobiont Bacteroides vulgatus activates NF-κB in a human gut epithelial cell line in a strain and growth phase dependent manner. Anaerobe. 2017; 47:209–217. PMID: 28583864.36. Shih CT, Yeh YT, Lin CC, Yang LY, Chiang CP. Akkermansia muciniphila is negatively correlated with hemoglobin A1c in refractory diabetes. Microorganisms. 2020; 8(9):1360.37. Ho LK, Tong VJ, Syn N, Nagarajan N, Tham EH, Tay SK, et al. Gut microbiota changes in children with autism spectrum disorder: a systematic review. Gut Pathog. 2020; 12(1):6. PMID: 32025243.
Article38. Magro DO, Santos A, Guadagnini D, de Godoy FM, Silva SH, Lemos WJ, et al. Remission in Crohn’s disease is accompanied by alterations in the gut microbiota and mucins production. Sci Rep. 2019; 9(1):13263. PMID: 31520001.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- Nutritional Support in Patients with Inflammatory Bowel Diseases
- Types of Special Infant Formulas Marketed in Korea and Their Indications
- A glycolipid adjuvant, 7DW8-5, provides a protective effect against colonic inflammation in mice by the recruitment of CD1d-restricted natural killer T cells
- Implication of Porphyromonas gingivalis in colitis and homeostasis of intestinal epithelium
- A Study of the Bystander Effect and Its Enhancement in HSV-TK Gene Therapy Using a Murine Neuroblastoma Model