Cancer Res Treat.
2019 Oct;51(4):1411-1419. 10.4143/crt.2018.663.
The Use of CD44 Variant 9 and Ki-67 Combination Can Predicts Prognosis Better Than Their Single Use in Early Gastric Cancer
- Affiliations
-
- 1Department of Internal Medicine, Institute of Health Sciences, Gyeongsang National University Changwon Hospital, Gyeongsang National University School of Medicine, Changwon, Korea.
- 2Department of Pathology, Institute of Health Sciences, Gyeongsang National University Hospital, Gyeongsang National University School of Medicine, Jinju, Korea.
- 3Department of Internal Medicine, Institute of Health Sciences, Gyeongsang National University Hospital, Gyeongsang National University School of Medicine, Jinju, Korea. lwshmo@hanmail.net
- 4Department of Surgery, Institute of Health Sciences, Gyeongsang National University Changwon Hospital, Gyeongsang National University School of Medicine, Changwon, Korea.
- 5Department of Surgery, Institute of Health Sciences, Gyeongsang National University Hospital, Gyeongsang National University School of Medicine, Jinju, Korea.
- KMID: 2460590
- DOI: http://doi.org/10.4143/crt.2018.663
Abstract
- PURPOSE
We previously demonstrated that CD44v9 and Ki-67 played an important role in predicting poor prognosis of early gastric cancer (EGC). However, little is known about combined use of both biomarkers as prognostic biomarker. The present study was performed to investigate the significance of CD44v9 and Ki-67 expression as a combination biomarker for EGC.
MATERIALS AND METHODS
With tissue microarray for 158 EGC tissues, we performed immunohistochemical staining for CD44v9 and Ki-67. The whole patients were divided into three groups (group A, CD44v9-negative/Ki-67-low; group B, neither group A or C; and group C, CD44v9-positive/Ki-67-high). Its clinical significance was re-analyzed with adjustment via propensity score matching (PSM). For validation, we performed bootstrap resampling.
RESULTS
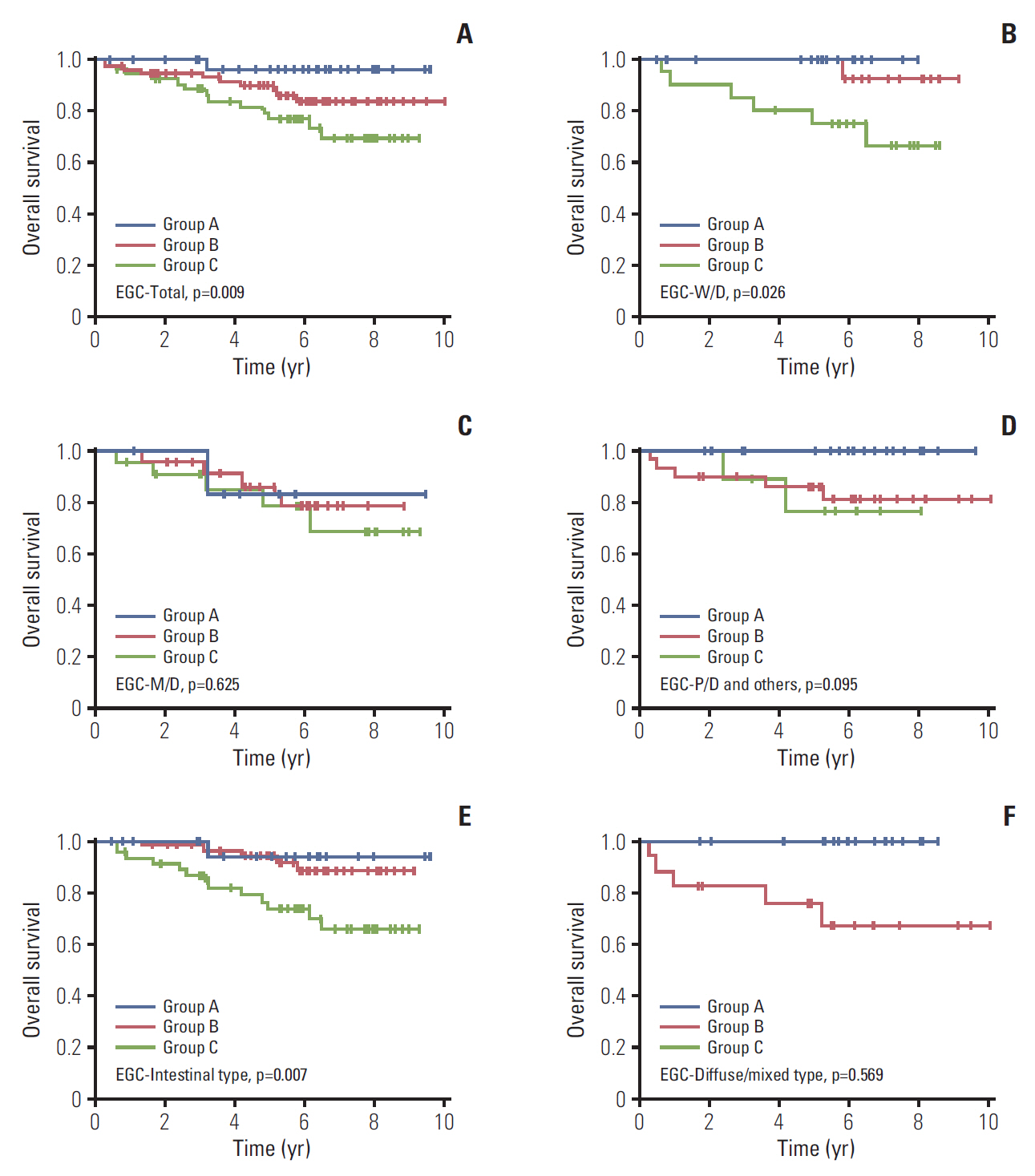
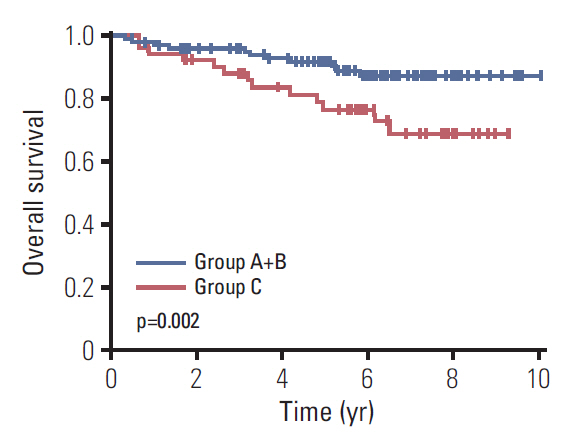
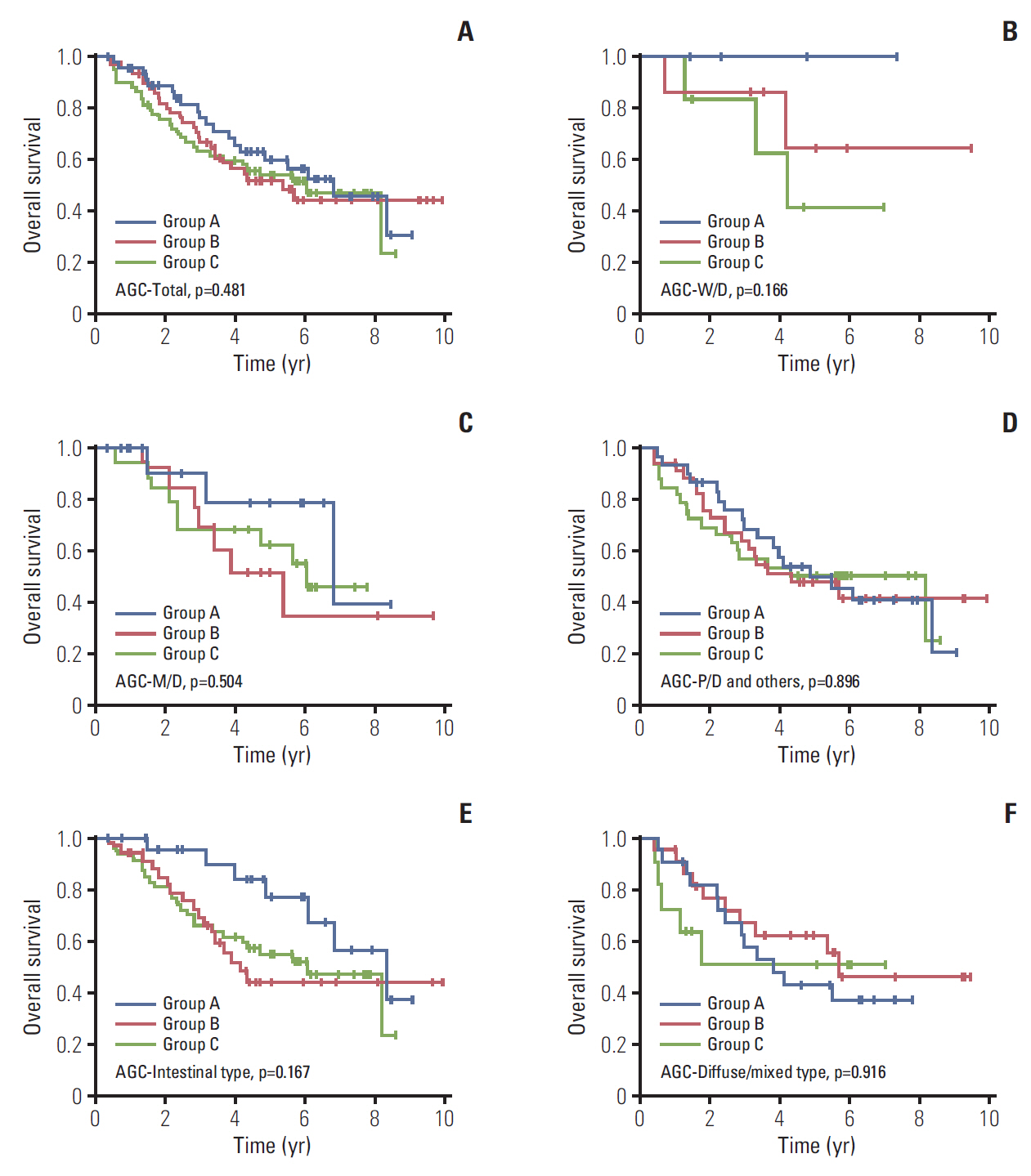
The median follow-up duration was 90.4 months (range, 3.7 to 120.4 months). In the comparison according to CD44v9/Ki-67 expression, the combined use of the two biomarker clearly separated the three groups by 5-year survival rates (5-YSR, 96.3%, 89.8%, and 76.8% in group A, B, and C, respectively; p=0.009). After PSM, 5-YSR were 97.7% and 76.8% in group A+B and group C, respectively (p=0.002). Multivariable analysis demonstrated that group C had independently poor prognosis (hazard ratio, 9.137; 95% confidence interval, 1.187 to 70.366; p=0.034) compared with group A. Bootstrap resampling internally validated this result (p=0.016).
CONCLUSION
This study suggests that both positive CD44v9 and high Ki-67 expression are associated with poor prognosis in EGC, and the combined use of these markers provides better prognostic stratification than the single use of them.
Keyword
MeSH Terms
Figure
Reference
-
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016; 66:7–30.
Article2. Song HN, Go SI, Lee WS, Kim Y, Choi HJ, Lee US, et al. Population-based regional cancer incidence in Korea: comparison between urban and rural areas. Cancer Res Treat. 2016; 48:789–97.
Article3. Oh CM, Won YJ, Jung KW, Kong HJ, Cho H, Lee JK, et al. Cancer statistics in Korea: incidence, mortality, survival, and prevalence in 2013. Cancer Res Treat. 2016; 48:436–50.
Article4. Ko GH, Go SI, Lee WS, Lee JH, Jeong SH, Lee YJ, et al. Prognostic impact of Ki-67 in patients with gastric cancer-the importance of depth of invasion and histologic differentiation. Medicine (Baltimore). 2017; 96:e7181.
Article5. Go SI, Ko GH, Lee WS, Kim RB, Lee JH, Jeong SH, et al. CD44 variant 9 serves as a poor prognostic marker in early gastric cancer, but not in advanced gastric cancer. Cancer Res Treat. 2016; 48:142–52.
Article6. Aruffo A, Stamenkovic I, Melnick M, Underhill CB, Seed B. CD44 is the principal cell surface receptor for hyaluronate. Cell. 1990; 61:1303–13.
Article7. Ishimoto T, Oshima H, Oshima M, Kai K, Torii R, Masuko T, et al. CD44+ slow-cycling tumor cell expansion is triggered by cooperative actions of Wnt and prostaglandin E2 in gastric tumorigenesis. Cancer Sci. 2010; 101:673–8.8. Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009; 458:780–3.
Article9. Culty M, Miyake K, Kincade PW, Sikorski E, Butcher EC, Underhill C. The hyaluronate receptor is a member of the CD44 (H-CAM) family of cell surface glycoproteins. J Cell Biol. 1990; 111(6 Pt 1):2765–74.10. Thomas L, Byers HR, Vink J, Stamenkovic I. CD44H regulates tumor cell migration on hyaluronate-coated substrate. J Cell Biol. 1992; 118:971–7.
Article11. Gunthert U, Hofmann M, Rudy W, Reber S, Zoller M, Haussmann I, et al. A new variant of glycoprotein CD44 confers metastatic potential to rat carcinoma cells. Cell. 1991; 65:13–24.
Article12. Ishimoto T, Nagano O, Yae T, Tamada M, Motohara T, Oshima H, et al. CD44 variant regulates redox status in cancer cells by stabilizing the xCT subunit of system xc(-) and thereby promotes tumor growth. Cancer Cell. 2011; 19:387–400.
Article13. Scholzen T, Gerdes J. The Ki-67 protein: from the known and the unknown. J Cell Physiol. 2000; 182:311–22.
Article14. Gerdes J, Lemke H, Baisch H, Wacker HH, Schwab U, Stein H. Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J Immunol. 1984; 133:1710–5.15. Deshmukh P, Ramsey L, Garewal HS. Ki-67 labeling index is a more reliable measure of solid tumor proliferative activity than tritiated thymidine labeling. Am J Clin Pathol. 1990; 94:192–5.
Article16. Cher ML, Chew K, Rosenau W, Carroll PR. Cellular proliferation in prostatic adenocarcinoma as assessed by bromodeoxyuridine uptake and Ki-67 and PCNA expression. Prostate. 1995; 26:87–93.
Article17. Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003; 100:3983–8.
Article18. Dalerba P, Dylla SJ, Park IK, Liu R, Wang X, Cho RW, et al. Phenotypic characterization of human colorectal cancer stem cells. Proc Natl Acad Sci U S A. 2007; 104:10158–63.
Article19. Zavros Y. Initiation and maintenance of gastric cancer: a focus on CD44 variant isoforms and cancer stem cells. Cell Mol Gastroenterol Hepatol. 2017; 4:55–63.
Article20. Singh SR. Gastric cancer stem cells: a novel therapeutic target. Cancer Lett. 2013; 338:110–9.
Article21. Kodama H, Murata S, Ishida M, Yamamoto H, Yamaguchi T, Kaida S, et al. Prognostic impact of CD44-positive cancer stem-like cells at the invasive front of gastric cancer. Br J Cancer. 2017; 116:186–94.
Article22. Luo G, Hu Y, Zhang Z, Wang P, Luo Z, Lin J, et al. Clinicopathologic significance and prognostic value of Ki-67 expression in patients with gastric cancer: a meta-analysis. Oncotarget. 2017; 8:50273–83.
Article23. Hu B, El Hajj N, Sittler S, Lammert N, Barnes R, Meloni-Ehrig A. Gastric cancer: classification, histology and application of molecular pathology. J Gastrointest Oncol. 2012; 3:251–61.24. Lauren P. The two histological main types of gastric carcinoma: diffuse and so-called intestinal-type carcinoma: an attempt at a histo-clinical classification. Acta Pathol Microbiol Scand. 1965; 64:31–49.25. Amin MB, Edge S, Greene F, Byrd DR, Brookland RK, Washington MK, et al. AJCC cancer staging manual. 8th ed. New York, NY: Springer;2016.26. Jerzak KJ, Pritchard KI. The 21-gene recurrence score assay in node-negative early breast cancer: prognostic, predictive or presumptuous? Eur J Cancer. 2016; 68:173–5.
Article27. Sestak I, Buus R, Cuzick J, Dubsky P, Kronenwett R, Denkert C, et al. Comparison of the performance of 6 prognostic signatures for estrogen receptor-positive breast cancer: a secondary analysis of a randomized clinical trial. JAMA Oncol. 2018; 4:545–53.28. Inoshita N, Yanagisawa A, Arai T, Kitagawa T, Hirokawa K, Kato Y. Pathological characteristics of gastric carcinomas in the very old. Jpn J Cancer Res. 1998; 89:1087–92.
Article29. Arai T, Esaki Y, Inoshita N, Sawabe M, Kasahara I, Kuroiwa K, et al. Pathologic characteristics of gastric cancer in the elderly: a retrospective study of 994 surgical patients. Gastric Cancer. 2004; 7:154–9.
Article30. Hassani Joutei HA, Marchoudi N, Sadaoui I, Mahfoud W, Haddad F, Fechtali T, et al. Ki-67 proliferative index in gastric cancer: a useful prognostic marker? Anticancer Res. 2014; 34:5806.31. Costa L, Fradique AC, Pupo A, Quaresma L, Cabrita F, Da Silva G, et al. Ki 67 showed statistical prognostic significance in intestinal type of gastric cancer. Ann Oncol. 2013; 24(Suppl 4):iv59.
Article32. Fang M, Wu J, Lai X, Ai H, Tao Y, Zhu B, et al. CD44 and CD44v6 are correlated with gastric cancer progression and poor patient prognosis: evidence from 42 studies. Cell Physiol Biochem. 2016; 40:567–78.
Article
- Full Text Links
- Actions
-
Cited
- CITED
-
- Close
- Share
- Similar articles
-
- The Serum Assay of Soluble CD44 Standard, CD44 Variant 5, and CD44 Variant 6 in Patients with Gastric Cancer
- The Role of CD44 in the Pathogenesis, Diagnosis, and Therapy of Gastric Cancer
- Expression of CD44 Variant 6 (V6) in endometrial cancer, endometrial hyperplasia, and normal endometrium
- CD44 Variant 9 Serves as a Poor Prognostic Marker in Early Gastric Cancer, But Not in Advanced Gastric Cancer
- Preferential Expression of CD44 in Thyroid Papillary Carcinoma